| functional annotation |
| Function |
CHASE domain containing histidine kinase protein |


|
| GO BP |
|
GO:0010086 [list] [network] embryonic root morphogenesis
|
(3 genes)
|
IMP
|
 |
|
GO:0007231 [list] [network] osmosensory signaling pathway
|
(5 genes)
|
IMP
|
 |
|
GO:0071329 [list] [network] cellular response to sucrose stimulus
|
(9 genes)
|
IMP
|
 |
|
GO:0008272 [list] [network] sulfate transport
|
(12 genes)
|
IMP
|
 |
|
GO:0033500 [list] [network] carbohydrate homeostasis
|
(27 genes)
|
IMP
|
 |
|
GO:0009116 [list] [network] nucleoside metabolic process
|
(62 genes)
|
IEA
|
 |
|
GO:0009736 [list] [network] cytokinin-activated signaling pathway
|
(66 genes)
|
TAS
|
 |
|
GO:0010029 [list] [network] regulation of seed germination
|
(84 genes)
|
IMP
|
 |
|
GO:0016036 [list] [network] cellular response to phosphate starvation
|
(110 genes)
|
IMP
|
 |
|
GO:0010150 [list] [network] leaf senescence
|
(111 genes)
|
IMP
|
 |
|
GO:0048509 [list] [network] regulation of meristem development
|
(125 genes)
|
IMP
|
 |
|
GO:0000160 [list] [network] phosphorelay signal transduction system
|
(234 genes)
|
IDA
|
 |
|
GO:0048831 [list] [network] regulation of shoot system development
|
(240 genes)
|
IMP
|
 |
|
GO:0009414 [list] [network] response to water deprivation
|
(361 genes)
|
IEP
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(399 genes)
|
IDA
IMP
|
 |
|
GO:0006468 [list] [network] protein phosphorylation
|
(921 genes)
|
IGI
|
 |
|
| GO CC |
|
GO:0005789 [list] [network] endoplasmic reticulum membrane
|
(466 genes)
|
IEA
|
 |
|
GO:0005783 [list] [network] endoplasmic reticulum
|
(877 genes)
|
IDA
|
 |
|
GO:0016021 [list] [network] integral component of membrane
|
(4803 genes)
|
IEA
|
 |
|
GO:0005634 [list] [network] nucleus
|
(10793 genes)
|
ISM
|
 |
|
| GO MF |
|
GO:0009885 [list] [network] transmembrane histidine kinase cytokinin receptor activity
|
(1 genes)
|
IDA
|
 |
|
GO:0009884 [list] [network] cytokinin receptor activity
|
(3 genes)
|
IDA
TAS
|
 |
|
GO:0005034 [list] [network] osmosensor activity
|
(4 genes)
|
IGI
|
 |
|
GO:0000155 [list] [network] phosphorelay sensor kinase activity
|
(16 genes)
|
IEA
|
 |
|
GO:0004673 [list] [network] protein histidine kinase activity
|
(17 genes)
|
IDA
ISS
|
 |
|
GO:0043424 [list] [network] protein histidine kinase binding
|
(22 genes)
|
IPI
|
 |
|
GO:0019901 [list] [network] protein kinase binding
|
(101 genes)
|
IPI
|
 |
|
GO:0004721 [list] [network] phosphoprotein phosphatase activity
|
(174 genes)
|
IDA
|
 |
|
GO:0019899 [list] [network] enzyme binding
|
(436 genes)
|
IPI
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
ath04075 [list] [network] Plant hormone signal transduction (273 genes) |
 |
| Protein |
NP_001324427.1
NP_001324428.1
NP_001324429.1
NP_565277.1
NP_849925.1
NP_973396.1
|
| BLAST |
NP_001324427.1
NP_001324428.1
NP_001324429.1
NP_565277.1
NP_849925.1
NP_973396.1
|
| Orthologous |
[Ortholog page]
LOC541634 (zma)
LOC732761 (zma)
LOC732762 (zma)
LOC732835 (zma)
LOC4333916 (osa)
LOC7457818 (ppo)
LOC7462655 (ppo)
LOC11411168 (mtr)
LOC100254304 (vvi)
LOC100776599 (gma)
LOC100789894 (gma)
LOC100813204 (gma)
LOC100818481 (gma)
LOC101262506 (sly)
LOC103827548 (bra)
LOC107275680 (osa)
|
Subcellular
localization
wolf |
|
plas 9,
nucl 1,
E.R. 1
|
(predict for NP_001324427.1)
|
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_001324428.1)
|
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_001324429.1)
|
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_565277.1)
|
|
plas 9,
E.R. 1
|
(predict for NP_849925.1)
|
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_973396.1)
|
|
Subcellular
localization
TargetP |
|
other 5,
chlo 3
|
(predict for NP_001324427.1)
|
|
other 9
|
(predict for NP_001324428.1)
|
|
other 9
|
(predict for NP_001324429.1)
|
|
other 9
|
(predict for NP_565277.1)
|
|
mito 5,
other 5
|
(predict for NP_849925.1)
|
|
other 9
|
(predict for NP_973396.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04075 |
Plant hormone signal transduction |
9 |
 |
| ath00908 |
Zeatin biosynthesis |
2 |
 |
Genes directly connected with WOL on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 7.8 |
RR3 |
response regulator 3 |
[detail] |
818730 |
| 6.9 |
ARR4 |
response regulator 4 |
[detail] |
837587 |
| 6.6 |
ARR6 |
response regulator 6 |
[detail] |
836412 |
| 5.5 |
AT2G40230 |
HXXXD-type acyl-transferase family protein |
[detail] |
818615 |
|
Coexpressed
gene list |
[Coexpressed gene list for WOL]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
263599_at
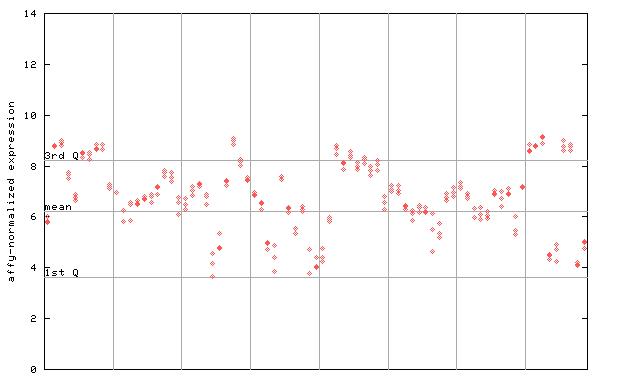
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
263599_at
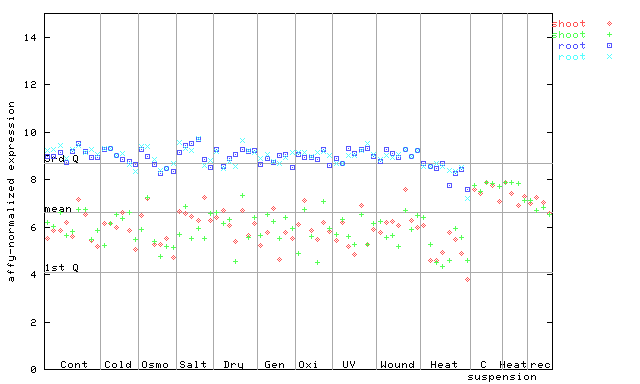
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
263599_at
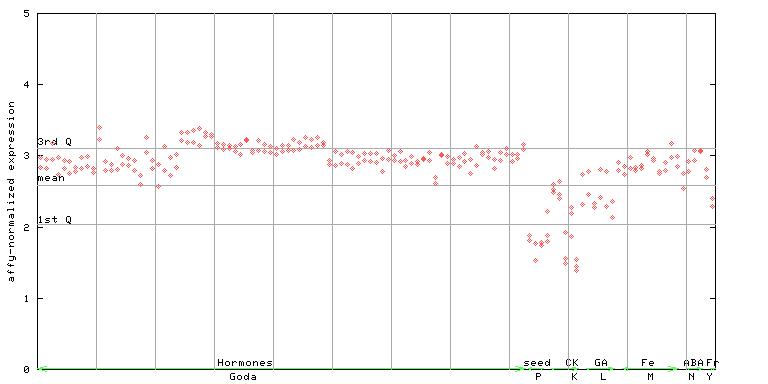
X axis is samples (xls file), and Y axis is log-expression.
|

















