| functional annotation |
| Function |
Leucine-rich repeat protein kinase family protein |


|
| GO BP |
|
GO:0048354 [list] [network] mucilage biosynthetic process involved in seed coat development
|
(14 genes)
|
IMP
|
 |
|
GO:0010192 [list] [network] mucilage biosynthetic process
|
(30 genes)
|
IMP
|
 |
|
GO:0009664 [list] [network] plant-type cell wall organization
|
(145 genes)
|
IMP
|
 |
|
GO:0046777 [list] [network] protein autophosphorylation
|
(173 genes)
|
IBA
|
 |
|
GO:0009826 [list] [network] unidimensional cell growth
|
(279 genes)
|
IMP
|
 |
|
| GO CC |
|
GO:0005576 [list] [network] extracellular region
|
(3363 genes)
|
ISM
|
 |
|
GO:0005886 [list] [network] plasma membrane
|
(3771 genes)
|
IBA
IDA
|
 |
|
GO:0016021 [list] [network] integral component of membrane
|
(4803 genes)
|
IEA
|
 |
|
| GO MF |
|
GO:0004674 [list] [network] protein serine/threonine kinase activity
|
(801 genes)
|
IBA
|
 |
|
GO:0016301 [list] [network] kinase activity
|
(1362 genes)
|
IDA
ISS
|
 |
|
GO:0005524 [list] [network] ATP binding
|
(2003 genes)
|
IEA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001189684.1
NP_001324112.1
NP_001324113.1
NP_181105.2
|
| BLAST |
NP_001189684.1
NP_001324112.1
NP_001324113.1
NP_181105.2
|
| Orthologous |
[Ortholog page]
FEI1 (ath)
LOC4332359 (osa)
LOC11410978 (mtr)
LOC100264793 (vvi)
LOC100280440 (zma)
LOC100281527 (zma)
LOC100816028 (gma)
LOC100819230 (gma)
LOC101257290 (sly)
LOC103857572 (bra)
LOC103865464 (bra)
LOC103867340 (bra)
|
Subcellular
localization
wolf |
|
chlo 3,
plas 2,
vacu 2,
extr 1,
chlo_mito 1,
nucl_plas 1,
E.R._vacu 1,
E.R._plas 1
|
(predict for NP_001189684.1)
|
|
chlo 3,
plas 2,
vacu 2,
extr 1,
chlo_mito 1,
nucl_plas 1,
E.R._vacu 1,
E.R._plas 1
|
(predict for NP_001324112.1)
|
|
chlo 3,
plas 2,
vacu 2,
extr 1,
chlo_mito 1,
nucl_plas 1,
E.R._vacu 1,
E.R._plas 1
|
(predict for NP_001324113.1)
|
|
chlo 3,
plas 2,
vacu 2,
extr 1,
chlo_mito 1,
nucl_plas 1,
E.R._vacu 1,
E.R._plas 1
|
(predict for NP_181105.2)
|
|
Subcellular
localization
TargetP |
|
scret 9
|
(predict for NP_001189684.1)
|
|
scret 9
|
(predict for NP_001324112.1)
|
|
scret 9
|
(predict for NP_001324113.1)
|
|
scret 9
|
(predict for NP_181105.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with FEI2 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 7.0 |
QUL1 |
QUASIMODO2 LIKE 1 |
[detail] |
837945 |
| 6.4 |
FEI1 |
Leucine-rich repeat protein kinase family protein |
[detail] |
840032 |
| 6.1 |
AT2G39750 |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
[detail] |
818560 |
| 6.0 |
AT4G38050 |
Xanthine/uracil permease family protein |
[detail] |
829961 |
| 5.1 |
AT3G60380 |
cotton fiber protein |
[detail] |
825209 |
|
Coexpressed
gene list |
[Coexpressed gene list for FEI2]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
265844_at
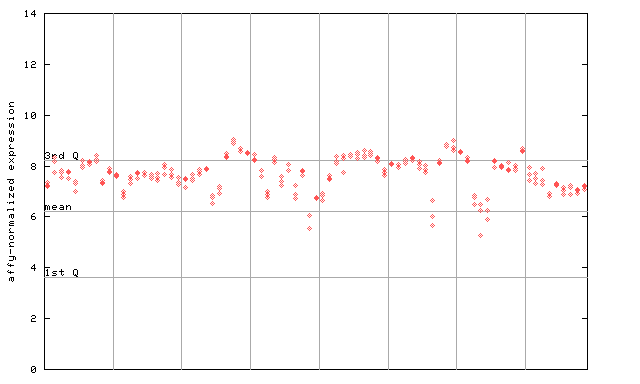
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
265844_at
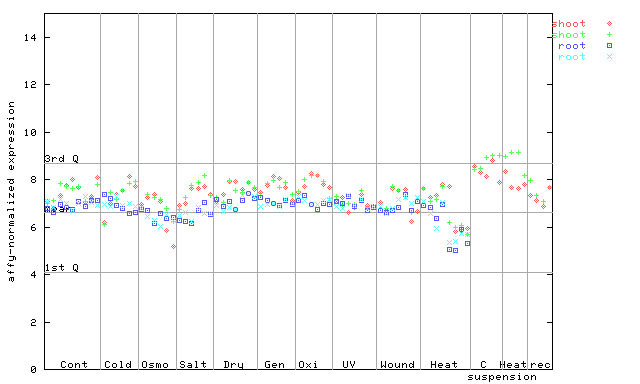
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
265844_at
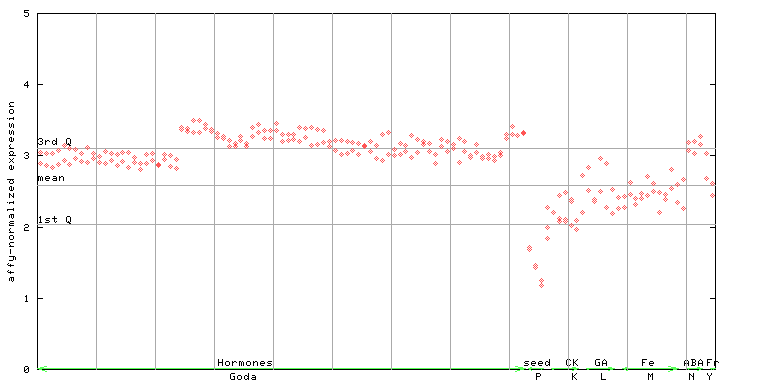
X axis is samples (xls file), and Y axis is log-expression.
|












