| functional annotation |
| Function |
SIGNAL PEPTIDE PEPTIDASE-LIKE 3 |


|
| GO BP |
|
GO:0033619 [list] [network] membrane protein proteolysis
|
(4 genes)
|
IBA
|
 |
|
| GO CC |
|
GO:0071556 [list] [network] integral component of lumenal side of endoplasmic reticulum membrane
|
(3 genes)
|
IBA
|
 |
|
GO:0071458 [list] [network] integral component of cytoplasmic side of endoplasmic reticulum membrane
|
(8 genes)
|
IBA
|
 |
|
GO:0005765 [list] [network] lysosomal membrane
|
(13 genes)
|
IBA
|
 |
|
GO:0030660 [list] [network] Golgi-associated vesicle membrane
|
(53 genes)
|
IBA
|
 |
|
GO:0010008 [list] [network] endosome membrane
|
(162 genes)
|
IEA
|
 |
|
GO:0005768 [list] [network] endosome
|
(424 genes)
|
TAS
|
 |
|
GO:0005774 [list] [network] vacuolar membrane
|
(624 genes)
|
IDA
|
 |
|
GO:0005886 [list] [network] plasma membrane
|
(3771 genes)
|
ISM
|
 |
|
| GO MF |
|
GO:0042500 [list] [network] aspartic endopeptidase activity, intramembrane cleaving
|
(3 genes)
|
IBA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001318409.1
NP_001325361.1
NP_001325362.1
NP_850383.3
|
| BLAST |
NP_001318409.1
NP_001325361.1
NP_001325362.1
NP_850383.3
|
| Orthologous |
[Ortholog page]
SPPL5 (ath)
LOC4350402 (osa)
LOC25485756 (mtr)
LOC25499789 (mtr)
LOC100217036 (zma)
LOC100249763 (vvi)
LOC100776679 (gma)
LOC100782203 (gma)
LOC100801070 (gma)
LOC100815578 (gma)
LOC101255676 (sly)
LOC101261270 (sly)
LOC103836510 (bra)
LOC103866691 (bra)
|
Subcellular
localization
wolf |
|
plas 9,
vacu 1,
golg 1
|
(predict for NP_001318409.1)
|
|
plas 8,
vacu 1,
E.R. 1,
golg 1,
E.R._vacu 1
|
(predict for NP_001325361.1)
|
|
plas 5,
vacu 2,
golg 2
|
(predict for NP_001325362.1)
|
|
plas 5,
vacu 1,
E.R. 1,
golg 1,
E.R._vacu 1
|
(predict for NP_850383.3)
|
|
Subcellular
localization
TargetP |
|
scret 9
|
(predict for NP_001318409.1)
|
|
scret 9
|
(predict for NP_001325361.1)
|
|
other 9
|
(predict for NP_001325362.1)
|
|
other 9
|
(predict for NP_850383.3)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00510 |
N-Glycan biosynthesis |
6 |
 |
| ath00513 |
Various types of N-glycan biosynthesis |
6 |
 |
| ath04141 |
Protein processing in endoplasmic reticulum |
6 |
 |
Genes directly connected with SPPL3 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 9.2 |
GPAT9 |
glycerol-3-phosphate acyltransferase 9 |
[detail] |
836183 |
| 8.2 |
KOB1 |
elongation defective 1 protein / ELD1 protein |
[detail] |
820002 |
| 7.7 |
HAP6 |
ribophorin II (RPN2) family protein |
[detail] |
827863 |
| 6.7 |
AT4G38790 |
ER lumen protein retaining receptor family protein |
[detail] |
830034 |
| 6.4 |
IAR1 |
ZIP metal ion transporter family |
[detail] |
843138 |
| 5.8 |
AT1G08750 |
Peptidase C13 family |
[detail] |
837397 |
| 5.8 |
AT3G14410 |
Nucleotide/sugar transporter family protein |
[detail] |
820663 |
| 4.9 |
PDE135 |
Xanthine/uracil permease family protein |
[detail] |
817192 |
| 4.5 |
AT3G06450 |
HCO3- transporter family |
[detail] |
819821 |
|
Coexpressed
gene list |
[Coexpressed gene list for SPPL3]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
266445_at
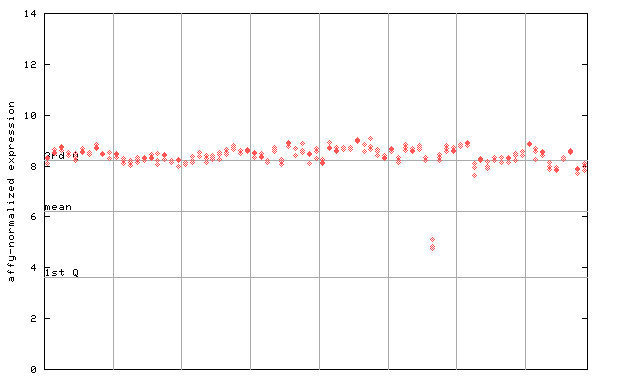
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
266445_at
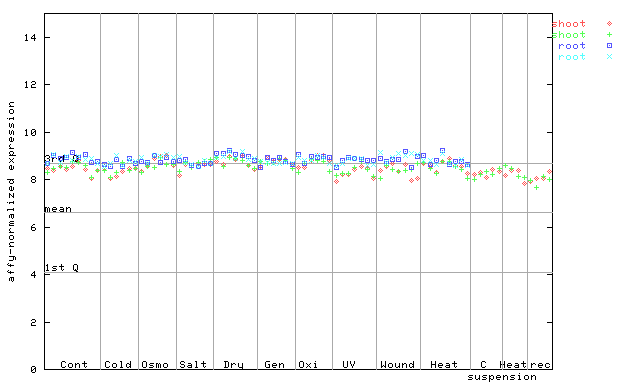
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
266445_at
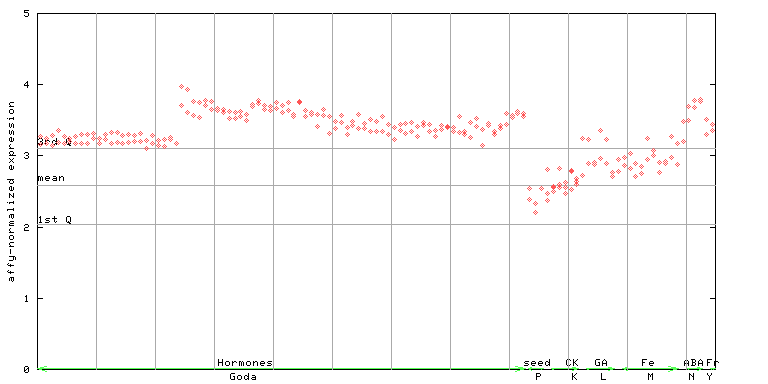
X axis is samples (xls file), and Y axis is log-expression.
|















