| functional annotation |
| Function |
glutamate receptor 1.1 |


|
| GO BP |
|
GO:0006814 [list] [network] sodium ion transport
|
(22 genes)
|
IDA
|
 |
|
GO:0006874 [list] [network] cellular calcium ion homeostasis
|
(51 genes)
|
NAS
|
 |
|
GO:0006816 [list] [network] calcium ion transport
|
(53 genes)
|
IDA
|
 |
|
GO:0006813 [list] [network] potassium ion transport
|
(81 genes)
|
IDA
|
 |
|
GO:0030003 [list] [network] cellular cation homeostasis
|
(156 genes)
|
IDA
|
 |
|
GO:0009738 [list] [network] abscisic acid-activated signaling pathway
|
(194 genes)
|
IEA
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(574 genes)
|
IMP
|
 |
|
GO:0009416 [list] [network] response to light stimulus
|
(700 genes)
|
IMP
|
 |
|
| GO CC |
|
GO:0005886 [list] [network] plasma membrane
|
(3771 genes)
|
IBA
ISM
|
 |
|
GO:0016021 [list] [network] integral component of membrane
|
(4803 genes)
|
IEA
|
 |
|
| GO MF |
|
GO:0005272 [list] [network] sodium channel activity
|
(1 genes)
|
IDA
|
 |
|
GO:0005262 [list] [network] calcium channel activity
|
(18 genes)
|
IDA
|
 |
|
GO:0005217 [list] [network] intracellular ligand-gated ion channel activity
|
(24 genes)
|
ISS
|
 |
|
GO:0005267 [list] [network] potassium channel activity
|
(26 genes)
|
IDA
|
 |
|
GO:0015276 [list] [network] ligand-gated ion channel activity
|
(33 genes)
|
IEA
|
 |
|
GO:0005261 [list] [network] cation channel activity
|
(75 genes)
|
IDA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001325545.1
NP_001325546.1
NP_187061.1
|
| BLAST |
NP_001325545.1
NP_001325546.1
NP_187061.1
|
| Orthologous |
[Ortholog page]
LOC103850056 (bra)
|
Subcellular
localization
wolf |
|
plas 6,
E.R. 1,
mito 1,
extr 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_001325545.1)
|
|
plas 5,
vacu 2,
E.R. 1,
mito 1,
extr 1,
cyto_E.R. 1
|
(predict for NP_001325546.1)
|
|
plas 6,
E.R. 1,
mito 1,
extr 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_187061.1)
|
|
Subcellular
localization
TargetP |
|
scret 9
|
(predict for NP_001325545.1)
|
|
scret 9
|
(predict for NP_001325546.1)
|
|
scret 9
|
(predict for NP_187061.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00460 |
Cyanoamino acid metabolism |
2 |
 |
Genes directly connected with GLR1.1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.1 |
AT1G51805 |
Leucine-rich repeat protein kinase family protein |
[detail] |
841607 |
| 5.4 |
AT5G39020 |
Malectin/receptor-like protein kinase family protein |
[detail] |
833894 |
| 5.2 |
CYP71B29 |
cytochrome P450, family 71, subfamily B, polypeptide 29 |
[detail] |
837867 |
| 4.8 |
MPK5 |
MAP kinase 5 |
[detail] |
826735 |
|
Coexpressed
gene list |
[Coexpressed gene list for GLR1.1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
258566_at
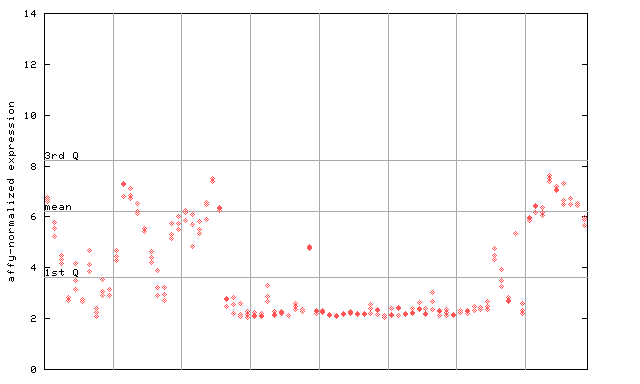
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
258566_at
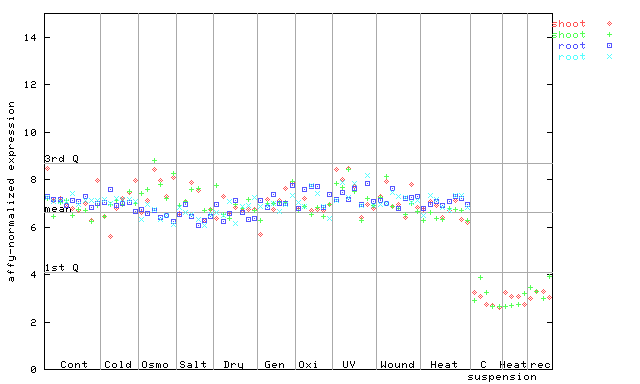
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
258566_at
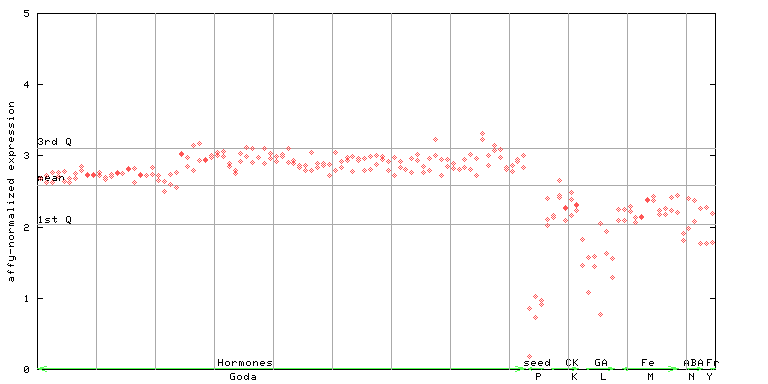
X axis is samples (xls file), and Y axis is log-expression.
|












