| functional annotation |
| Function |
Paxneb protein-like protein |


|
| GO BP |
|
GO:0043609 [list] [network] regulation of carbon utilization
|
(2 genes)
|
IMP
|
 |
|
GO:0031538 [list] [network] negative regulation of anthocyanin metabolic process
|
(3 genes)
|
IMP
|
 |
|
GO:0071329 [list] [network] cellular response to sucrose stimulus
|
(9 genes)
|
IMP
|
 |
|
GO:0035265 [list] [network] organ growth
|
(12 genes)
|
IMP
|
 |
|
GO:0002098 [list] [network] tRNA wobble uridine modification
|
(15 genes)
|
IEA
|
 |
|
GO:0008284 [list] [network] positive regulation of cell population proliferation
|
(26 genes)
|
IMP
|
 |
|
GO:0010928 [list] [network] regulation of auxin mediated signaling pathway
|
(30 genes)
|
IMP
|
 |
|
GO:2000024 [list] [network] regulation of leaf development
|
(73 genes)
|
IMP
|
 |
|
GO:0009738 [list] [network] abscisic acid-activated signaling pathway
|
(194 genes)
|
IEA
|
 |
|
GO:0009734 [list] [network] auxin-activated signaling pathway
|
(196 genes)
|
IEA
|
 |
|
GO:0051301 [list] [network] cell division
|
(352 genes)
|
IMP
|
 |
|
GO:0006979 [list] [network] response to oxidative stress
|
(442 genes)
|
IMP
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(574 genes)
|
IMP
|
 |
|
GO:0007275 [list] [network] multicellular organism development
|
(2714 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0016746 [list] [network] transferase activity, transferring acyl groups
|
(395 genes)
|
IEA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001319522.1
NP_566388.2
|
| BLAST |
NP_001319522.1
NP_566388.2
|
| Orthologous |
[Ortholog page]
LOC4341622 (osa)
LOC25500105 (mtr)
LOC100192820 (zma)
LOC100257804 (vvi)
LOC100799237 (gma)
LOC101253042 (sly)
LOC103870366 (bra)
|
Subcellular
localization
wolf |
|
nucl 6,
mito 2,
chlo_mito 2,
chlo 1
|
(predict for NP_001319522.1)
|
|
chlo 6,
chlo_mito 5,
mito 2
|
(predict for NP_566388.2)
|
|
Subcellular
localization
TargetP |
|
chlo 5
|
(predict for NP_001319522.1)
|
|
chlo 5
|
(predict for NP_566388.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with ELO1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 5.5 |
ATE2 |
arginine-tRNA protein transferase 2 |
[detail] |
820295 |
| 5.1 |
AT5G60590 |
DHBP synthase RibB-like alpha/beta domain-containing protein |
[detail] |
836180 |
| 4.7 |
AT3G21140 |
Pyridoxamine 5'-phosphate oxidase family protein |
[detail] |
821666 |
|
Coexpressed
gene list |
[Coexpressed gene list for ELO1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
256412_at
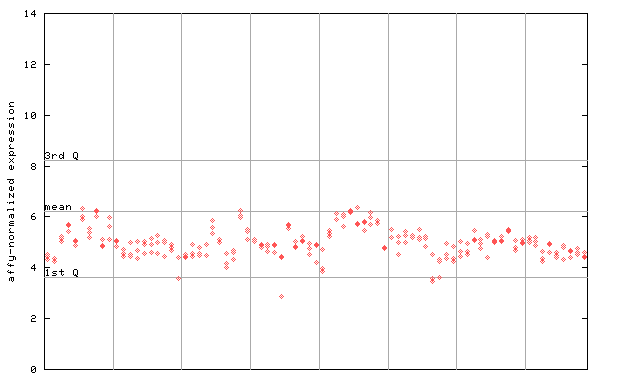
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
256412_at
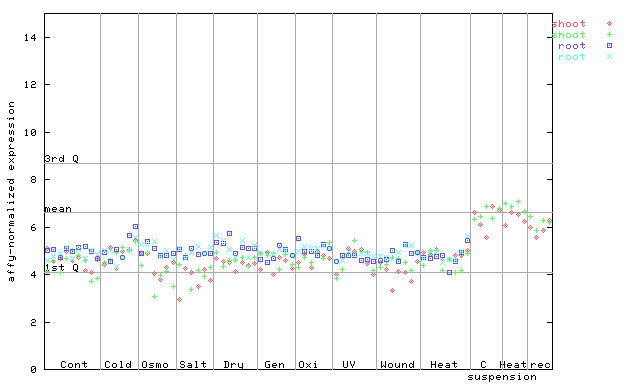
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
256412_at
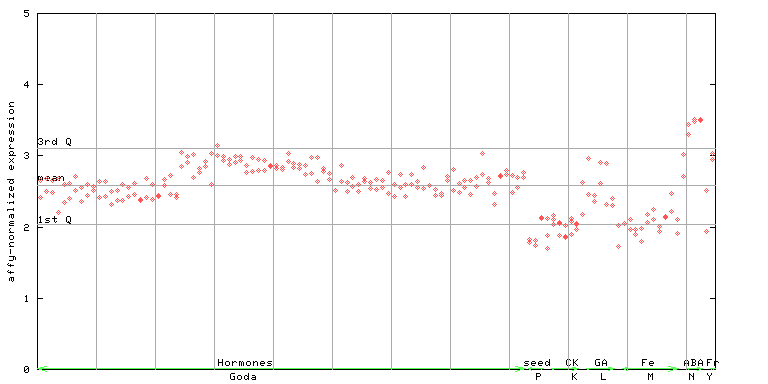
X axis is samples (xls file), and Y axis is log-expression.
|










