[←][→] ath
| functional annotation | ||||||||||||||||||||||||||
| Function | Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A protein |


|
||||||||||||||||||||||||
| GO BP |
|
|||||||||||||||||||||||||
| GO CC |
|
|||||||||||||||||||||||||
| GO MF |
|
|||||||||||||||||||||||||
| KEGG | ||||||||||||||||||||||||||
| Protein | NP_188110.1 | |||||||||||||||||||||||||
| BLAST | NP_188110.1 | |||||||||||||||||||||||||
| Orthologous | [Ortholog page] LOC4327096 (osa) LOC7472347 (ppo) LOC11419220 (mtr) LOC25482180 (mtr) LOC100246994 (vvi) LOC100253845 (vvi) LOC100316898 (sly) LOC100783515 (gma) LOC100785850 (gma) LOC101260612 (sly) LOC103634961 (zma) LOC103841763 (bra) LOC103859598 (bra) LOC112937639 (osa) | |||||||||||||||||||||||||
| Subcellular localization wolf |
|
|||||||||||||||||||||||||
| Subcellular localization TargetP |
|
|||||||||||||||||||||||||
| Gene coexpression | ||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for AT3G14920] | |||||||||||||||||||||||||
| Gene expression | ||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | |||||||||||||||||||||||||
| AtGenExpress* (Development) |
257209_at
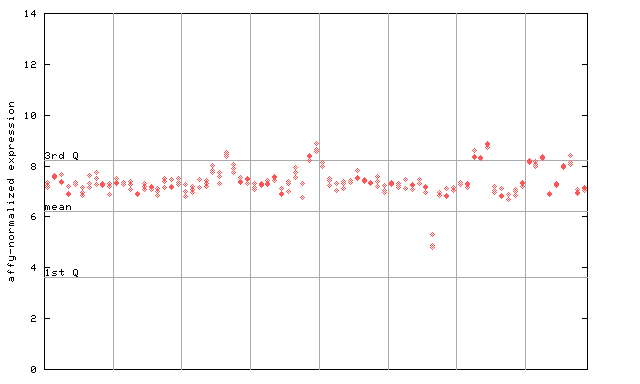
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||
| AtGenExpress* (Stress) |
257209_at
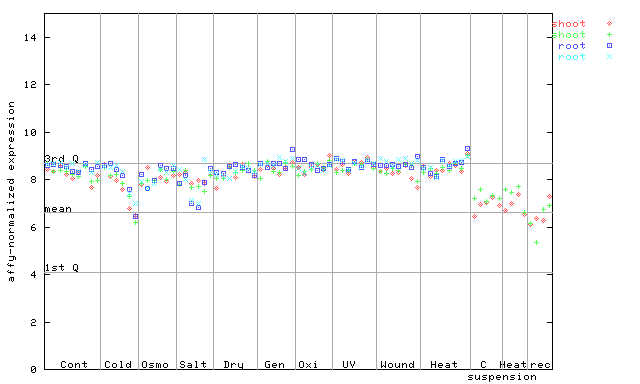
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
257209_at
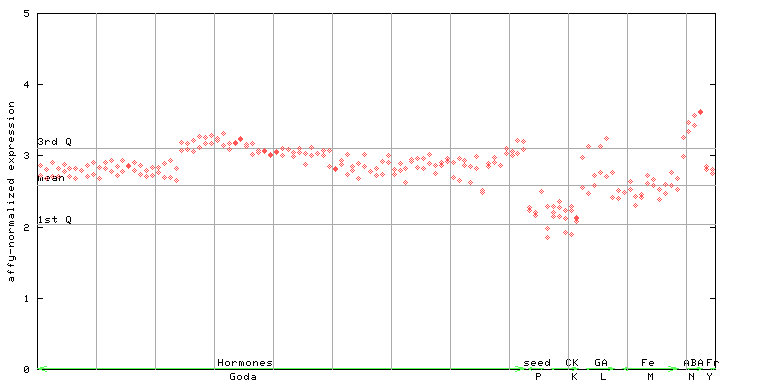
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 820721 |


|
| Refseq ID (protein) | NP_188110.1 |  |
The preparation time of this page was 0.2 [sec].

