| functional annotation |
| Function |
SNF1 kinase homolog 10 |


|
| GO BP |
|
GO:0009594 [list] [network] detection of nutrient
|
(1 genes)
|
IDA
|
 |
|
GO:0010508 [list] [network] positive regulation of autophagy
|
(8 genes)
|
IMP
|
 |
|
GO:0010050 [list] [network] vegetative phase change
|
(10 genes)
|
IMP
|
 |
|
GO:0009635 [list] [network] response to herbicide
|
(13 genes)
|
IEP
|
 |
|
GO:1902074 [list] [network] response to salt
|
(20 genes)
|
IEP
|
 |
|
GO:0080022 [list] [network] primary root development
|
(32 genes)
|
IMP
|
 |
|
GO:1900055 [list] [network] regulation of leaf senescence
|
(36 genes)
|
IDA
|
 |
|
GO:0010182 [list] [network] sugar mediated signaling pathway
|
(37 genes)
|
IMP
|
 |
|
GO:0009789 [list] [network] positive regulation of abscisic acid-activated signaling pathway
|
(40 genes)
|
IMP
|
 |
|
GO:0042128 [list] [network] nitrate assimilation
|
(41 genes)
|
IEA
|
 |
|
GO:0005982 [list] [network] starch metabolic process
|
(59 genes)
|
IMP
|
 |
|
GO:0009749 [list] [network] response to glucose
|
(59 genes)
|
IDA
|
 |
|
GO:0010150 [list] [network] leaf senescence
|
(111 genes)
|
IMP
|
 |
|
GO:0006633 [list] [network] fatty acid biosynthetic process
|
(150 genes)
|
IEA
|
 |
|
GO:0010228 [list] [network] vegetative to reproductive phase transition of meristem
|
(173 genes)
|
IMP
|
 |
|
GO:0009738 [list] [network] abscisic acid-activated signaling pathway
|
(194 genes)
|
IMP
|
 |
|
GO:0001666 [list] [network] response to hypoxia
|
(263 genes)
|
IDA
|
 |
|
GO:0006979 [list] [network] response to oxidative stress
|
(442 genes)
|
IEP
|
 |
|
GO:0035556 [list] [network] intracellular signal transduction
|
(517 genes)
|
IBA
|
 |
|
GO:0006468 [list] [network] protein phosphorylation
|
(921 genes)
|
IBA
|
 |
|
GO:0099402 [list] [network] plant organ development
|
(981 genes)
|
IMP
|
 |
|
GO:0003006 [list] [network] developmental process involved in reproduction
|
(1475 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001118546.1
NP_566130.1
NP_850488.1
|
| BLAST |
NP_001118546.1
NP_566130.1
NP_850488.1
|
| Orthologous |
[Ortholog page]
LOC542687 (zma)
LOC543870 (sly)
SNRK1 (gma)
SNF1 (sly)
KIN11 (ath)
SnRK1.3 (ath)
LOC4332495 (osa)
LOC4339410 (osa)
LOC4342639 (osa)
LOC4345873 (osa)
LOC7480582 (ppo)
LOC7497360 (ppo)
LOC11412507 (mtr)
LOC25496433 (mtr)
LOC100254223 (vvi)
LOC100384241 (zma)
LOC100796687 (gma)
SNRK1.1 (gma)
LOC103639865 (zma)
LOC103647175 (zma)
LOC103842817 (bra)
LOC103854153 (bra)
LOC103858930 (bra)
LOC103863845 (bra)
LOC103870947 (bra)
LOC103875101 (bra)
|
Subcellular
localization
wolf |
|
cyto 5,
nucl 2,
chlo 1,
vacu 1,
E.R. 1,
golg 1,
E.R._vacu 1
|
(predict for NP_001118546.1)
|
|
cyto 5,
nucl 2,
chlo 1,
vacu 1,
E.R. 1,
golg 1,
E.R._vacu 1
|
(predict for NP_566130.1)
|
|
chlo 2,
cyto 2,
golg 2,
nucl 1,
golg_plas 1,
cyto_E.R. 1,
cyto_plas 1
|
(predict for NP_850488.1)
|
|
Subcellular
localization
TargetP |
|
other 8
|
(predict for NP_001118546.1)
|
|
other 8
|
(predict for NP_566130.1)
|
|
other 7
|
(predict for NP_850488.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04141 |
Protein processing in endoplasmic reticulum |
2 |
 |
Genes directly connected with KIN10 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.5 |
KIN11 |
SNF1 kinase homolog 11 |
[detail] |
822566 |
| 6.4 |
SNF4 |
sucrose nonfermenting-like protein |
[detail] |
837423 |
| 6.3 |
AT2G28390 |
SAND family protein |
[detail] |
817387 |
| 4.9 |
STY8 |
ACT-like protein tyrosine kinase family protein |
[detail] |
816278 |
|
Coexpressed
gene list |
[Coexpressed gene list for KIN10]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
259319_at
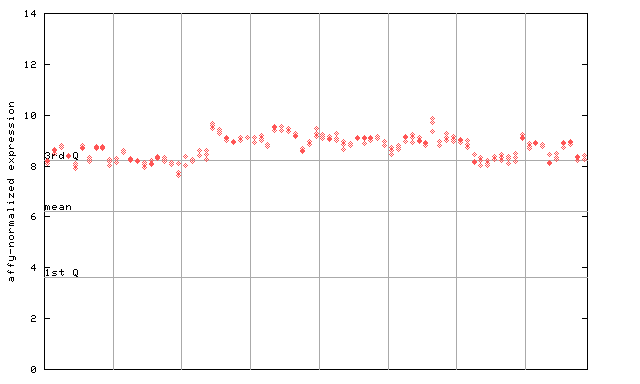
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
259319_at
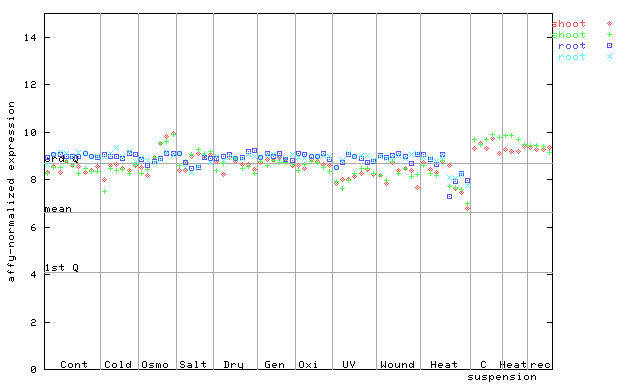
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
259319_at
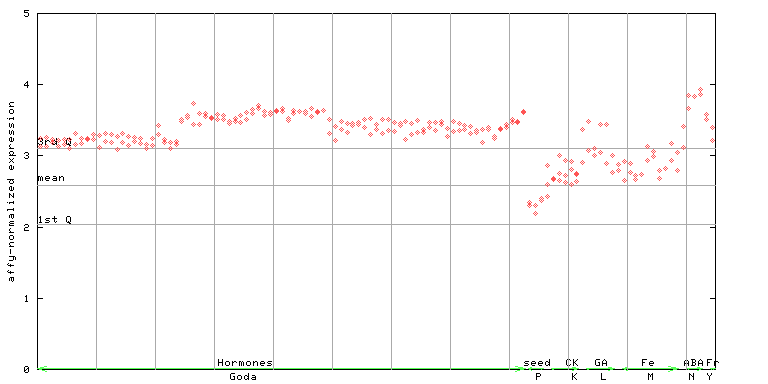
X axis is samples (xls file), and Y axis is log-expression.
|












