| functional annotation |
| Function |
MutT/nudix family protein |


|
| GO BP |
|
GO:0070212 [list] [network] protein poly-ADP-ribosylation
|
(4 genes)
|
IDA
|
 |
|
GO:0010581 [list] [network] regulation of starch biosynthetic process
|
(8 genes)
|
IMP
|
 |
|
GO:0009870 [list] [network] defense response signaling pathway, resistance gene-dependent
|
(15 genes)
|
IMP
|
 |
|
GO:0010193 [list] [network] response to ozone
|
(36 genes)
|
IEP
|
 |
|
GO:0009626 [list] [network] plant-type hypersensitive response
|
(73 genes)
|
IMP
|
 |
|
GO:0071456 [list] [network] cellular response to hypoxia
|
(238 genes)
|
HEP
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(399 genes)
|
IMP
|
 |
|
GO:0006979 [list] [network] response to oxidative stress
|
(442 genes)
|
IEP
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(485 genes)
|
IEP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0080042 [list] [network] ADP-glucose pyrophosphohydrolase activity
|
(2 genes)
|
IDA
|
 |
|
GO:0080041 [list] [network] ADP-ribose pyrophosphohydrolase activity
|
(3 genes)
|
IDA
|
 |
|
GO:0017110 [list] [network] nucleoside-diphosphatase activity
|
(4 genes)
|
IDA
|
 |
|
GO:0000210 [list] [network] NAD+ diphosphatase activity
|
(8 genes)
|
IDA
IEA
|
 |
|
GO:0047631 [list] [network] ADP-ribose diphosphatase activity
|
(10 genes)
|
IDA
|
 |
|
GO:0051287 [list] [network] NAD binding
|
(66 genes)
|
IDA
|
 |
|
GO:0046872 [list] [network] metal ion binding
|
(3180 genes)
|
IEA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001190707.1
NP_001329573.1
NP_001329574.1
NP_193008.1
NP_849367.1
NP_849368.1
|
| BLAST |
NP_001190707.1
NP_001329573.1
NP_001329574.1
NP_193008.1
NP_849367.1
NP_849368.1
|
| Orthologous |
[Ortholog page]
NUDT5 (ath)
NUDT6 (ath)
NUDT10 (ath)
NUDT2 (ath)
LOC4341595 (osa)
LOC7456012 (ppo)
LOC7463650 (ppo)
LOC7471520 (ppo)
LOC7482637 (ppo)
LOC7487546 (ppo)
LOC7491923 (ppo)
LOC25486266 (mtr)
LOC25490725 (mtr)
LOC25494321 (mtr)
LOC100192595 (zma)
LOC100253608 (vvi)
LOC100256580 (vvi)
LOC100257937 (vvi)
LOC100259630 (vvi)
LOC100273780 (zma)
LOC100778567 (gma)
LOC100783941 (gma)
LOC100810351 (gma)
LOC100815518 (gma)
LOC100818821 (gma)
LOC100819062 (gma)
LOC101245949 (sly)
LOC101259154 (sly)
LOC101260552 (sly)
LOC101263765 (sly)
LOC101267872 (sly)
LOC103827629 (bra)
LOC103833538 (bra)
LOC103839513 (bra)
LOC103853829 (bra)
LOC103860821 (bra)
LOC103863229 (bra)
LOC109123568 (vvi)
|
Subcellular
localization
wolf |
|
cyto 6,
nucl 1,
cysk 1,
cysk_nucl 1
|
(predict for NP_001190707.1)
|
|
cyto 4,
nucl 4,
chlo 1
|
(predict for NP_001329573.1)
|
|
cyto 5,
pero 2,
chlo 1,
extr 1,
cysk 1,
golg 1
|
(predict for NP_001329574.1)
|
|
cysk 4,
cyto 4,
nucl 1
|
(predict for NP_193008.1)
|
|
cysk 4,
cyto 4,
nucl 1
|
(predict for NP_849367.1)
|
|
cysk 4,
cyto 4,
nucl 1
|
(predict for NP_849368.1)
|
|
Subcellular
localization
TargetP |
|
other 8
|
(predict for NP_001190707.1)
|
|
other 7
|
(predict for NP_001329573.1)
|
|
other 8
|
(predict for NP_001329574.1)
|
|
other 8
|
(predict for NP_193008.1)
|
|
other 8
|
(predict for NP_849367.1)
|
|
other 8
|
(predict for NP_849368.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04626 |
Plant-pathogen interaction |
4 |
 |
Genes directly connected with NUDT7 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 13.3 |
TCH3 |
Calcium-binding EF hand family protein |
[detail] |
818709 |
| 11.4 |
GILP |
GSH-induced LITAF domain protein |
[detail] |
831158 |
| 11.3 |
ACA1 |
autoinhibited Ca2+-ATPase 1 |
[detail] |
839670 |
| 9.9 |
CPK28 |
calcium-dependent protein kinase 28 |
[detail] |
836753 |
| 9.6 |
SYP121 |
syntaxin of plants 121 |
[detail] |
820355 |
| 9.5 |
AT4G34150 |
Calcium-dependent lipid-binding (CaLB domain) family protein |
[detail] |
829563 |
| 9.2 |
AT1G69840 |
SPFH/Band 7/PHB domain-containing membrane-associated protein family |
[detail] |
843320 |
| 8.3 |
CPK4 |
calcium-dependent protein kinase 4 |
[detail] |
826541 |
| 8.1 |
AT2G20142 |
Toll-Interleukin-Resistance (TIR) domain family protein |
[detail] |
2745551 |
| 7.2 |
S6K2 |
serine/threonine protein kinase 2 |
[detail] |
820019 |
| 7.0 |
CNGC19 |
cyclic nucleotide gated channel 19 |
[detail] |
821036 |
| 6.8 |
AT1G09932 |
Phosphoglycerate mutase family protein |
[detail] |
837526 |
| 6.8 |
VPS60.1 |
SNF7 family protein |
[detail] |
820233 |
| 6.0 |
AT2G43320 |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
[detail] |
818933 |
| 5.6 |
AT5G41180 |
leucine-rich repeat transmembrane protein kinase family protein |
[detail] |
834120 |
|
Coexpressed
gene list |
[Coexpressed gene list for NUDT7]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
254784_at
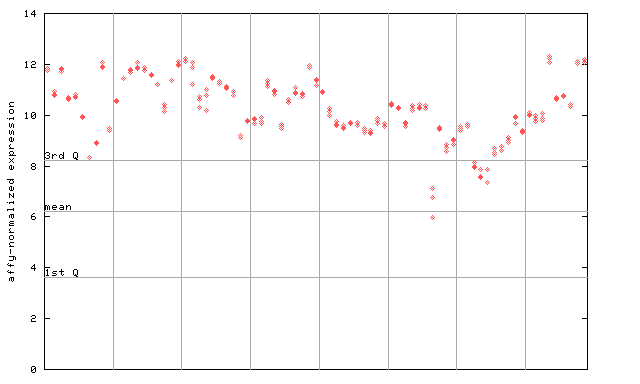
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
254784_at
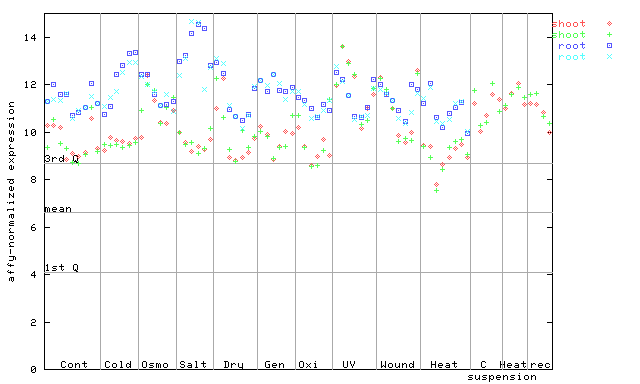
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
254784_at
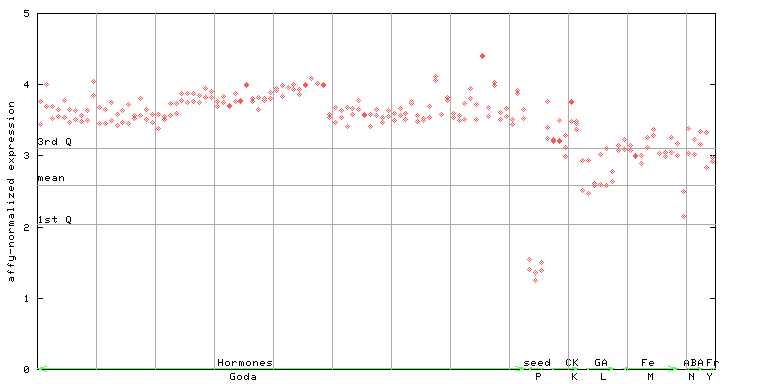
X axis is samples (xls file), and Y axis is log-expression.
|















