| functional annotation |
| Function |
separase |


|
| GO BP |
|
GO:0045876 [list] [network] positive regulation of sister chromatid cohesion
|
(3 genes)
|
IMP
|
 |
|
GO:2000114 [list] [network] regulation of establishment of cell polarity
|
(5 genes)
|
IMP
|
 |
|
GO:0051307 [list] [network] meiotic chromosome separation
|
(12 genes)
|
IBA
IMP
|
 |
|
GO:0051304 [list] [network] chromosome separation
|
(14 genes)
|
IMP
|
 |
|
GO:0009960 [list] [network] endosperm development
|
(34 genes)
|
IMP
|
 |
|
GO:0006887 [list] [network] exocytosis
|
(53 genes)
|
IMP
|
 |
|
GO:0009749 [list] [network] response to glucose
|
(59 genes)
|
IMP
|
 |
|
GO:0000911 [list] [network] cytokinesis by cell plate formation
|
(63 genes)
|
IMP
|
 |
|
GO:0009826 [list] [network] unidimensional cell growth
|
(279 genes)
|
IMP
|
 |
|
GO:0016192 [list] [network] vesicle-mediated transport
|
(417 genes)
|
IMP
|
 |
|
GO:0009793 [list] [network] embryo development ending in seed dormancy
|
(547 genes)
|
IMP
|
 |
|
GO:0006508 [list] [network] proteolysis
|
(901 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001190804.1
NP_001328112.1
NP_194028.2
|
| BLAST |
NP_001190804.1
NP_001328112.1
NP_194028.2
|
| Orthologous |
[Ortholog page]
LOC4330866 (osa)
LOC11418825 (mtr)
LOC25497699 (mtr)
LOC100259948 (vvi)
LOC100791010 (gma)
LOC100805306 (gma)
LOC101263947 (sly)
LOC103654220 (zma)
LOC103860637 (bra)
LOC103861751 (bra)
|
Subcellular
localization
wolf |
|
nucl 5,
plas 2,
cyto 1,
cysk_plas 1,
mito_plas 1
|
(predict for NP_001190804.1)
|
|
nucl 5,
plas 2,
cyto 1,
cysk_plas 1,
mito_plas 1
|
(predict for NP_001328112.1)
|
|
nucl 5,
plas 2,
cyto 1,
cysk_plas 1,
mito_plas 1
|
(predict for NP_194028.2)
|
|
Subcellular
localization
TargetP |
|
other 5,
chlo 3
|
(predict for NP_001190804.1)
|
|
other 5,
chlo 3
|
(predict for NP_001328112.1)
|
|
other 5,
chlo 3
|
(predict for NP_194028.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03030 |
DNA replication |
3 |
 |
| ath03440 |
Homologous recombination |
3 |
 |
| ath03410 |
Base excision repair |
2 |
 |
| ath03420 |
Nucleotide excision repair |
2 |
 |
Genes directly connected with ESP on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 7.8 |
TIL1 |
DNA polymerase epsilon catalytic subunit |
[detail] |
837346 |
| 7.5 |
AT4G14970 |
fanconi anemia group D2 protein |
[detail] |
827156 |
| 7.5 |
AT3G10180 |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
820180 |
| 6.3 |
AT3G20490 |
Rho GTPase-activating protein |
[detail] |
821595 |
| 5.7 |
PC-MYB1 |
Homeodomain-like protein |
[detail] |
829409 |
|
Coexpressed
gene list |
[Coexpressed gene list for ESP]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
254288_at
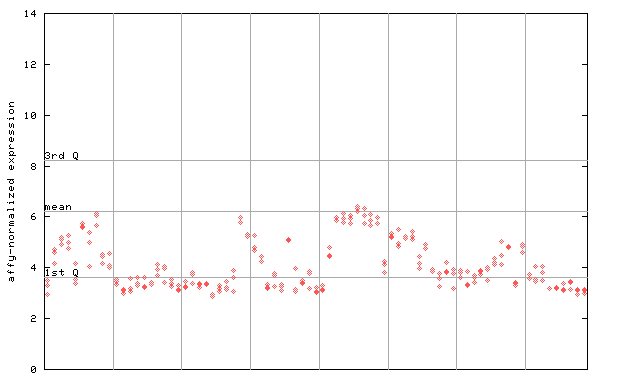
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
254288_at
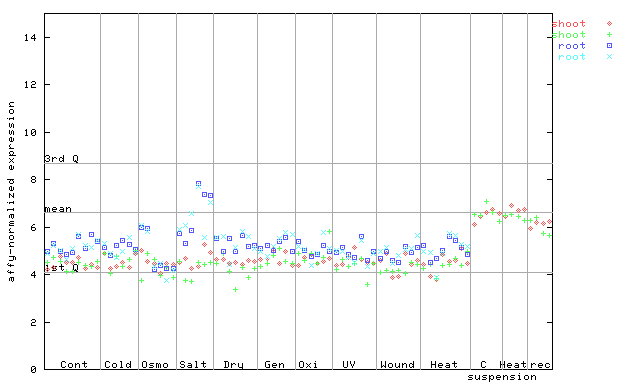
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
254288_at
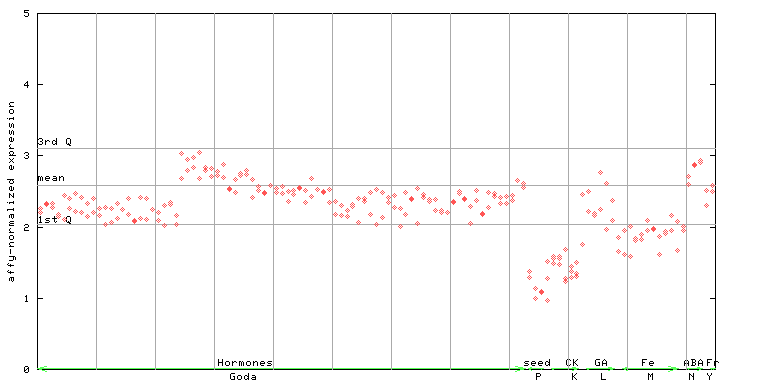
X axis is samples (xls file), and Y axis is log-expression.
|















