| functional annotation |
| Function |
protein CTF7 |


|
| GO BP |
|
GO:0034089 [list] [network] establishment of meiotic sister chromatid cohesion
|
(1 genes)
|
IMP
|
 |
|
GO:0034421 [list] [network] post-translational protein acetylation
|
(1 genes)
|
IBA
|
 |
|
GO:0060772 [list] [network] leaf phyllotactic patterning
|
(4 genes)
|
IMP
|
 |
|
GO:0080186 [list] [network] developmental vegetative growth
|
(10 genes)
|
IMP
|
 |
|
GO:0000070 [list] [network] mitotic sister chromatid segregation
|
(24 genes)
|
IMP
|
 |
|
GO:0007062 [list] [network] sister chromatid cohesion
|
(35 genes)
|
IBA
IMP
|
 |
|
GO:0006275 [list] [network] regulation of DNA replication
|
(49 genes)
|
IBA
|
 |
|
GO:0045132 [list] [network] meiotic chromosome segregation
|
(56 genes)
|
IMP
|
 |
|
GO:0000724 [list] [network] double-strand break repair via homologous recombination
|
(68 genes)
|
IMP
|
 |
|
GO:0048653 [list] [network] anther development
|
(75 genes)
|
IMP
|
 |
|
GO:0009553 [list] [network] embryo sac development
|
(146 genes)
|
IMP
|
 |
|
GO:0048609 [list] [network] multicellular organismal reproductive process
|
(173 genes)
|
IMP
|
 |
|
GO:0051301 [list] [network] cell division
|
(352 genes)
|
IEA
|
 |
|
GO:0048364 [list] [network] root development
|
(482 genes)
|
IMP
|
 |
|
GO:0009793 [list] [network] embryo development ending in seed dormancy
|
(547 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0016407 [list] [network] acetyltransferase activity
|
(141 genes)
|
IDA
|
 |
|
GO:0046872 [list] [network] metal ion binding
|
(3180 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001328055.1
NP_194868.2
|
| BLAST |
NP_001328055.1
NP_194868.2
|
| Orthologous |
[Ortholog page]
LOC4336297 (osa)
LOC4338615 (osa)
LOC7469952 (ppo)
LOC11432514 (mtr)
LOC25479864 (mtr)
LOC100242830 (vvi)
LOC100249805 (vvi)
LOC100280348 (zma)
LOC100809303 (gma)
LOC100811441 (gma)
LOC101248340 (sly)
LOC101260855 (sly)
LOC103646265 (zma)
LOC103648542 (zma)
LOC103834613 (bra)
LOC103862051 (bra)
|
Subcellular
localization
wolf |
|
nucl 6,
chlo 2,
cyto 1,
chlo_mito 1
|
(predict for NP_001328055.1)
|
|
nucl 9,
chlo 1,
cyto 1
|
(predict for NP_194868.2)
|
|
Subcellular
localization
TargetP |
|
scret 3,
chlo 3
|
(predict for NP_001328055.1)
|
|
scret 3,
chlo 3
|
(predict for NP_194868.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03030 |
DNA replication |
4 |
 |
| ath03440 |
Homologous recombination |
4 |
 |
| ath03420 |
Nucleotide excision repair |
3 |
 |
| ath03430 |
Mismatch repair |
3 |
 |
Genes directly connected with CTF7 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 7.1 |
CYCA3;1 |
Cyclin A3;1 |
[detail] |
834324 |
| 6.8 |
RPA70D |
Replication factor-A protein 1-like protein |
[detail] |
836221 |
| 6.7 |
RPA70B |
RPA70-kDa subunit B |
[detail] |
830696 |
|
Coexpressed
gene list |
[Coexpressed gene list for CTF7]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
253518_at
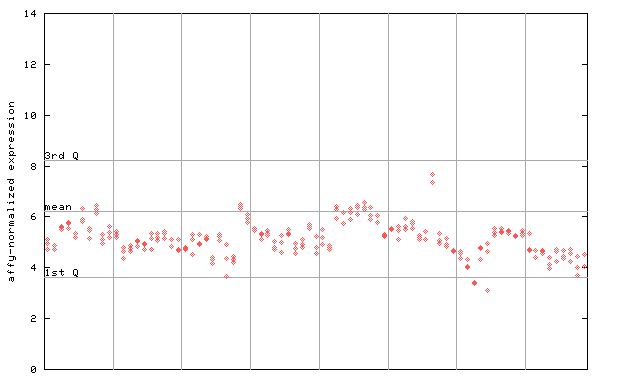
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
253518_at
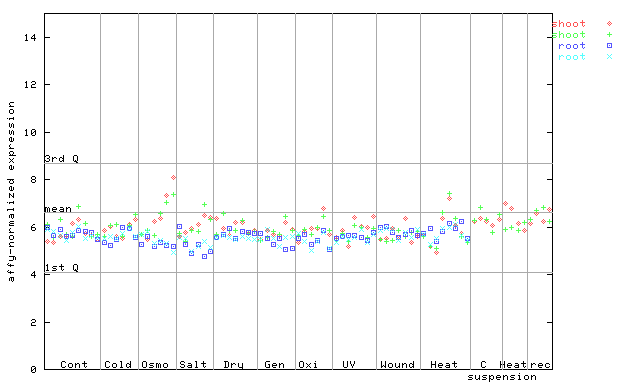
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
253518_at
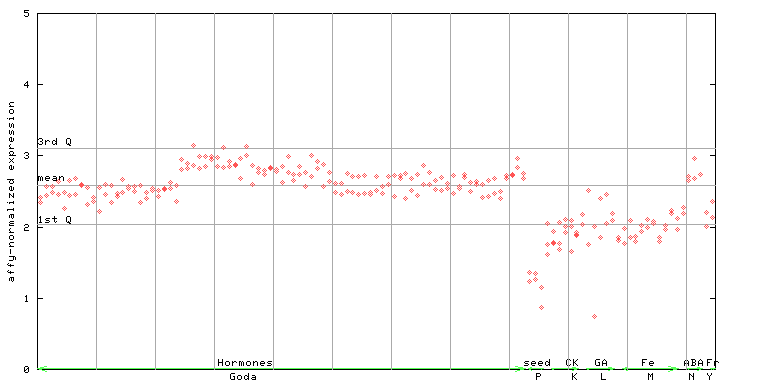
X axis is samples (xls file), and Y axis is log-expression.
|














