| functional annotation |
| Function |
NRAMP metal ion transporter family protein |


|
| GO BP |
|
GO:0001736 [list] [network] establishment of planar polarity
|
(3 genes)
|
IGI
|
 |
|
GO:0009871 [list] [network] jasmonic acid and ethylene-dependent systemic resistance, ethylene mediated signaling pathway
|
(4 genes)
|
TAS
|
 |
|
GO:0052544 [list] [network] defense response by callose deposition in cell wall
|
(16 genes)
|
IMP
|
 |
|
GO:0002237 [list] [network] response to molecule of bacterial origin
|
(34 genes)
|
IMP
|
 |
|
GO:0010182 [list] [network] sugar mediated signaling pathway
|
(37 genes)
|
TAS
|
 |
|
GO:0009789 [list] [network] positive regulation of abscisic acid-activated signaling pathway
|
(40 genes)
|
IMP
|
 |
|
GO:0031348 [list] [network] negative regulation of defense response
|
(63 genes)
|
IMP
|
 |
|
GO:0009736 [list] [network] cytokinin-activated signaling pathway
|
(66 genes)
|
IEA
|
 |
|
GO:0009926 [list] [network] auxin polar transport
|
(66 genes)
|
IMP
|
 |
|
GO:0010119 [list] [network] regulation of stomatal movement
|
(88 genes)
|
IMP
|
 |
|
GO:0010087 [list] [network] phloem or xylem histogenesis
|
(102 genes)
|
IMP
|
 |
|
GO:0048765 [list] [network] root hair cell differentiation
|
(109 genes)
|
IGI
|
 |
|
GO:0010150 [list] [network] leaf senescence
|
(111 genes)
|
IMP
|
 |
|
GO:0008219 [list] [network] cell death
|
(127 genes)
|
NAS
|
 |
|
GO:0009873 [list] [network] ethylene-activated signaling pathway
|
(191 genes)
|
TAS
|
 |
|
GO:0009734 [list] [network] auxin-activated signaling pathway
|
(196 genes)
|
IEA
|
 |
|
GO:0009753 [list] [network] response to jasmonic acid
|
(222 genes)
|
IMP
|
 |
|
GO:0009408 [list] [network] response to heat
|
(229 genes)
|
IMP
|
 |
|
GO:0009723 [list] [network] response to ethylene
|
(297 genes)
|
IMP
|
 |
|
GO:0051301 [list] [network] cell division
|
(352 genes)
|
IMP
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(399 genes)
|
IMP
|
 |
|
GO:0006979 [list] [network] response to oxidative stress
|
(442 genes)
|
TAS
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(485 genes)
|
IMP
|
 |
|
GO:0050832 [list] [network] defense response to fungus
|
(517 genes)
|
IMP
|
 |
|
GO:0006970 [list] [network] response to osmotic stress
|
(563 genes)
|
IMP
|
 |
|
GO:0009725 [list] [network] response to hormone
|
(1564 genes)
|
IMP
|
 |
|
| GO CC |
|
GO:0005789 [list] [network] endoplasmic reticulum membrane
|
(466 genes)
|
IEA
|
 |
|
GO:0016021 [list] [network] integral component of membrane
|
(4803 genes)
|
IEA
|
 |
|
GO:0005634 [list] [network] nucleus
|
(10793 genes)
|
IEA
|
 |
|
| GO MF |
|
GO:0015086 [list] [network] cadmium ion transmembrane transporter activity
|
(17 genes)
|
IBA
|
 |
|
GO:0005384 [list] [network] manganese ion transmembrane transporter activity
|
(25 genes)
|
IBA
|
 |
|
GO:0003729 [list] [network] mRNA binding
|
(1026 genes)
|
IDA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
ath04016 [list] [network] MAPK signaling pathway - plant (134 genes) |
 |
| ath04075 [list] [network] Plant hormone signal transduction (273 genes) |
 |
| Protein |
NP_195948.1
|
| BLAST |
NP_195948.1
|
| Orthologous |
[Ortholog page]
EIN2 (sly)
LOC4333824 (osa)
LOC4342431 (osa)
LOC4342433 (osa)
LOC7467627 (ppo)
LOC25479910 (mtr)
LOC100256742 (vvi)
LOC100781309 (gma)
LOC100801190 (gma)
LOC100813034 (gma)
LOC103649560 (zma)
LOC103847394 (bra)
LOC107278000 (osa)
|
Subcellular
localization
wolf |
|
plas 4,
vacu 1,
E.R. 1,
E.R._vacu 1,
chlo 1,
cyto 1,
mito 1,
extr 1,
chlo_mito 1
|
(predict for NP_195948.1)
|
|
Subcellular
localization
TargetP |
|
other 4,
scret 3
|
(predict for NP_195948.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04144 |
Endocytosis |
3 |
 |
| ath00190 |
Oxidative phosphorylation |
2 |
 |
Genes directly connected with EIN2 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 7.7 |
AT5G15680 |
ARM repeat superfamily protein |
[detail] |
831422 |
| 7.1 |
EDA10 |
SEC7-like guanine nucleotide exchange family protein |
[detail] |
839301 |
| 6.9 |
AT2G35050 |
kinase superfamily with octicosapeptide/Phox/Bem1p domain-containing protein |
[detail] |
818070 |
| 6.2 |
HA1 |
H[+]-ATPase 1 |
[detail] |
816413 |
| 5.1 |
AT1G34110 |
Leucine-rich receptor-like protein kinase family protein |
[detail] |
840310 |
|
Coexpressed
gene list |
[Coexpressed gene list for EIN2]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
250928_at
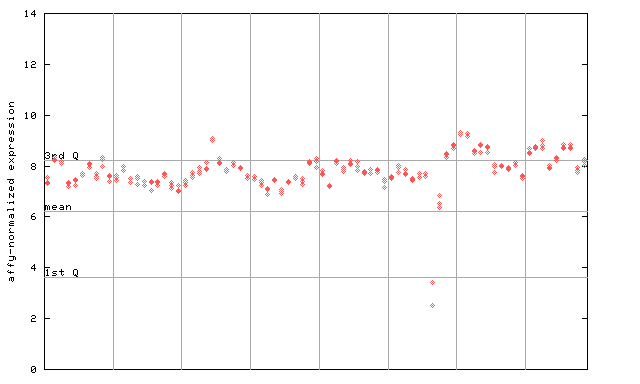
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
250928_at
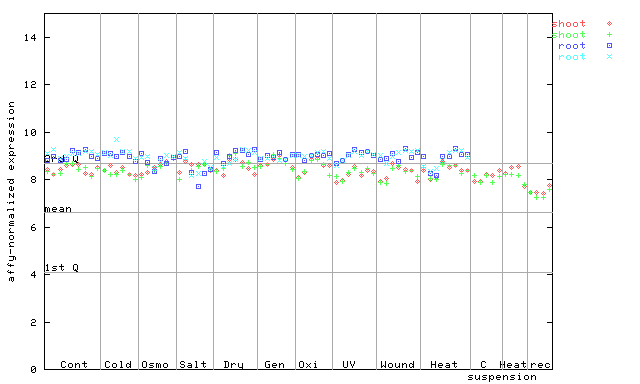
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
250928_at
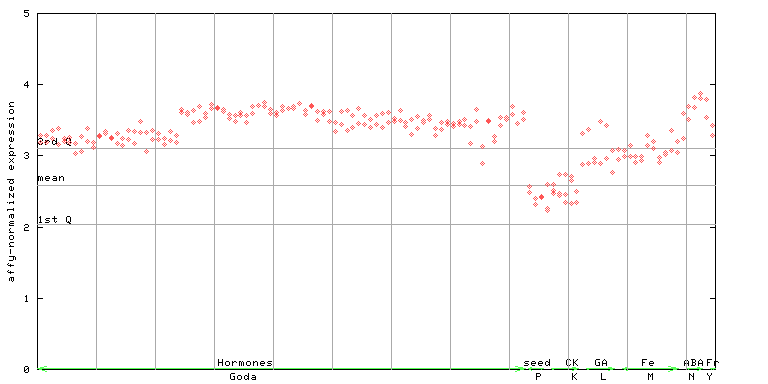
X axis is samples (xls file), and Y axis is log-expression.
|













