[←][→] ath
| functional annotation | |||||||||||||||||||||||||||||||||||||||
| Function | chromatin remodeling 42 |


|
|||||||||||||||||||||||||||||||||||||
| GO BP |
|
||||||||||||||||||||||||||||||||||||||
| GO CC |
|
||||||||||||||||||||||||||||||||||||||
| GO MF |
|
||||||||||||||||||||||||||||||||||||||
| KEGG | ath03440 [list] [network] Homologous recombination (67 genes) |  |
|||||||||||||||||||||||||||||||||||||
| Protein | NP_197542.1 | ||||||||||||||||||||||||||||||||||||||
| BLAST | NP_197542.1 | ||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] CHR38 (ath) LOC4344376 (osa) LOC7462098 (ppo) LOC11408204 (mtr) LOC11435042 (mtr) LOC11441869 (mtr) LOC25484660 (mtr) LOC100250037 (vvi) LOC100781856 (gma) LOC100785036 (gma) LOC100811703 (gma) LOC101262122 (sly) LOC103633601 (zma) LOC103834007 (bra) LOC103845677 (bra) | ||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
||||||||||||||||||||||||||||||||||||||
| Gene coexpression | |||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for CHR42] | ||||||||||||||||||||||||||||||||||||||
| Gene expression | |||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | ||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
246077_at
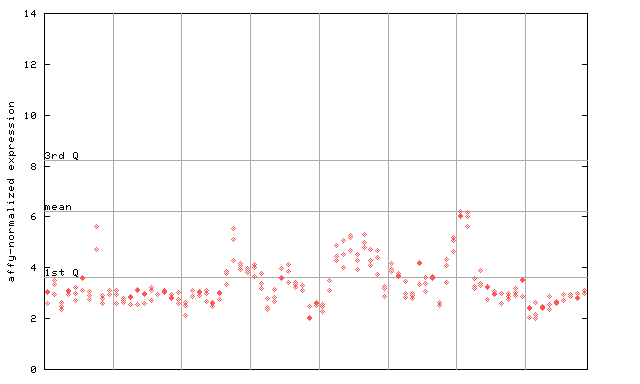
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
246077_at
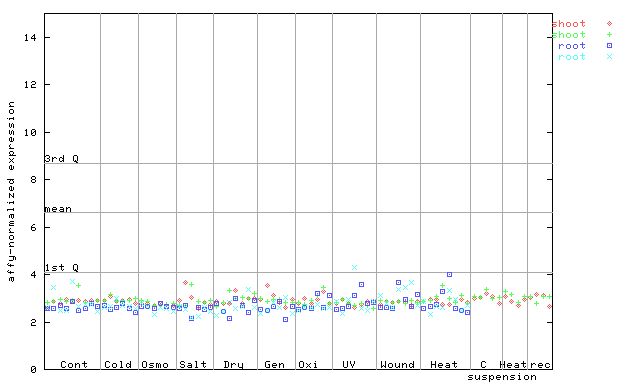
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
246077_at
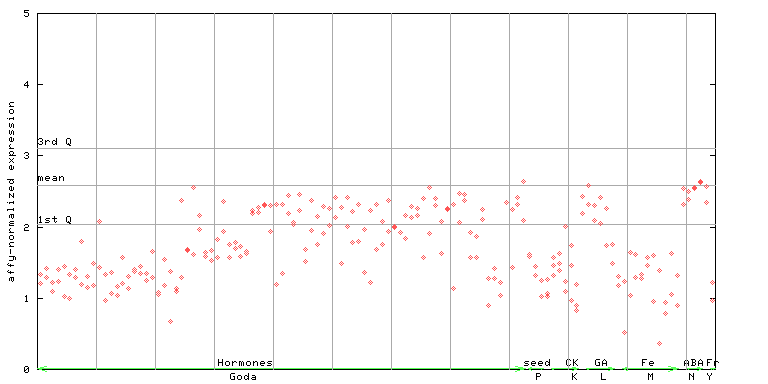
X axis is samples (xls file), and Y axis is log-expression. |
||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 832164 |


|
| Refseq ID (protein) | NP_197542.1 |  |
The preparation time of this page was 0.2 [sec].


