| functional annotation |
| Function |
RAS associated with diabetes protein 51 |


|
| GO BP |
|
GO:1990426 [list] [network] mitotic recombination-dependent replication fork processing
|
(1 genes)
|
IEA
|
 |
|
GO:0000730 [list] [network] DNA recombinase assembly
|
(2 genes)
|
IBA
|
 |
|
GO:0042148 [list] [network] strand invasion
|
(2 genes)
|
IBA
|
 |
|
GO:0045003 [list] [network] double-strand break repair via synthesis-dependent strand annealing
|
(7 genes)
|
IDA
|
 |
|
GO:0006312 [list] [network] mitotic recombination
|
(10 genes)
|
IBA
|
 |
|
GO:0010332 [list] [network] response to gamma radiation
|
(16 genes)
|
IEP
|
 |
|
GO:0007131 [list] [network] reciprocal meiotic recombination
|
(49 genes)
|
IBA
|
 |
|
GO:0070192 [list] [network] chromosome organization involved in meiotic cell cycle
|
(50 genes)
|
IBA
|
 |
|
GO:0006302 [list] [network] double-strand break repair
|
(108 genes)
|
IMP
|
 |
|
GO:0006259 [list] [network] DNA metabolic process
|
(441 genes)
|
IDA
TAS
|
 |
|
GO:0009314 [list] [network] response to radiation
|
(723 genes)
|
NAS
|
 |
|
GO:0006355 [list] [network] regulation of transcription, DNA-templated
|
(1984 genes)
|
IDA
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
ath03440 [list] [network] Homologous recombination (67 genes) |
 |
| Protein |
NP_001332342.1
NP_568402.1
|
| BLAST |
NP_001332342.1
NP_568402.1
|
| Orthologous |
[Ortholog page]
LOC541709 (zma)
LOC541710 (zma)
RAD51 (sly)
LOC4350923 (osa)
LOC4352257 (osa)
LOC11418607 (mtr)
LOC100252629 (vvi)
LOC100812213 (gma)
LOC100820307 (gma)
LOC103845639 (bra)
LOC103856468 (bra)
|
Subcellular
localization
wolf |
|
cyto 5,
chlo 1,
pero 1,
nucl 1,
mito 1,
plas 1,
nucl_plas 1,
mito_plas 1
|
(predict for NP_001332342.1)
|
|
cyto 5,
pero 2,
nucl 1,
mito 1,
plas 1,
cysk_nucl 1,
mito_plas 1
|
(predict for NP_568402.1)
|
|
Subcellular
localization
TargetP |
|
other 8,
mito 4
|
(predict for NP_001332342.1)
|
|
other 8,
mito 4
|
(predict for NP_568402.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03440 |
Homologous recombination |
6 |
 |
| ath03030 |
DNA replication |
3 |
 |
| ath03420 |
Nucleotide excision repair |
3 |
 |
| ath03430 |
Mismatch repair |
3 |
 |
| ath00240 |
Pyrimidine metabolism |
2 |
 |
Genes directly connected with RAD51 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 13.4 |
BRCA1 |
breast cancer susceptibility1 |
[detail] |
827854 |
| 10.9 |
AT4G02110 |
transcription coactivator |
[detail] |
828135 |
| 10.8 |
WEE1 |
WEE1-like kinase |
[detail] |
839453 |
| 10.5 |
GR1 |
protein gamma response 1 |
[detail] |
824375 |
| 8.6 |
AT4G19130 |
Replication factor-A protein 1-like protein |
[detail] |
827651 |
| 7.8 |
TK1a |
Thymidine kinase |
[detail] |
819971 |
| 5.4 |
UNG |
uracil dna glycosylase |
[detail] |
821394 |
|
Coexpressed
gene list |
[Coexpressed gene list for RAD51]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
246132_at
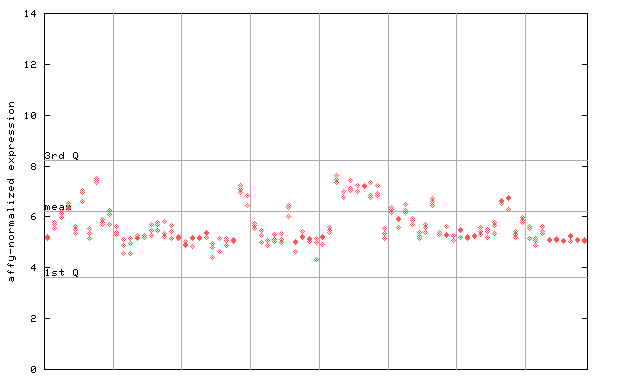
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
246132_at
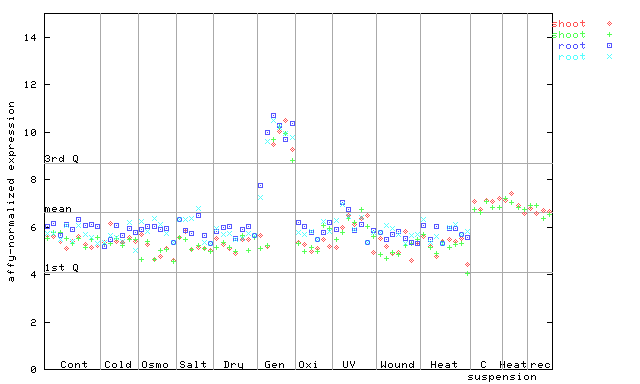
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
246132_at
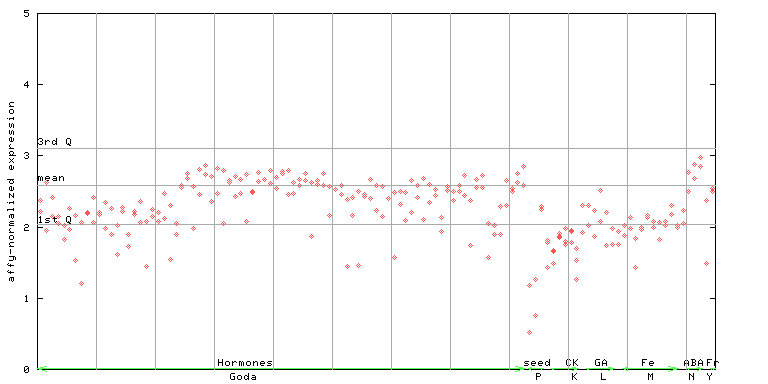
X axis is samples (xls file), and Y axis is log-expression.
|
















