| functional annotation |
| Function |
Ataxia telangiectasia-mutated and RAD3-like protein |


|
| GO BP |
|
GO:0043247 [list] [network] telomere maintenance in response to DNA damage
|
(6 genes)
|
TAS
|
 |
|
GO:0000077 [list] [network] DNA damage checkpoint
|
(10 genes)
|
IBA
|
 |
|
GO:0032204 [list] [network] regulation of telomere maintenance
|
(10 genes)
|
IGI
|
 |
|
GO:0007004 [list] [network] telomere maintenance via telomerase
|
(11 genes)
|
IMP
|
 |
|
GO:0010044 [list] [network] response to aluminum ion
|
(14 genes)
|
IMP
|
 |
|
GO:0006303 [list] [network] double-strand break repair via nonhomologous end joining
|
(16 genes)
|
IMP
|
 |
|
GO:0010332 [list] [network] response to gamma radiation
|
(16 genes)
|
IMP
|
 |
|
GO:0006282 [list] [network] regulation of DNA repair
|
(29 genes)
|
IMP
|
 |
|
GO:0000723 [list] [network] telomere maintenance
|
(36 genes)
|
IBA
|
 |
|
GO:0033044 [list] [network] regulation of chromosome organization
|
(99 genes)
|
IGI
|
 |
|
GO:0051321 [list] [network] meiotic cell cycle
|
(166 genes)
|
IGI
|
 |
|
GO:0032504 [list] [network] multicellular organism reproduction
|
(180 genes)
|
IGI
|
 |
|
GO:0031347 [list] [network] regulation of defense response
|
(260 genes)
|
IMP
|
 |
|
GO:0006281 [list] [network] DNA repair
|
(295 genes)
|
IGI
|
 |
|
GO:0006952 [list] [network] defense response
|
(1420 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0004674 [list] [network] protein serine/threonine kinase activity
|
(801 genes)
|
IBA
|
 |
|
GO:0005524 [list] [network] ATP binding
|
(2003 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001332642.1
NP_001332643.1
NP_001332644.1
NP_198898.2
|
| BLAST |
NP_001332642.1
NP_001332643.1
NP_001332644.1
NP_198898.2
|
| Orthologous |
[Ortholog page]
LOC4342103 (osa)
LOC7470423 (ppo)
LOC11440718 (mtr)
LOC100245131 (vvi)
LOC100804117 (gma)
LOC101256634 (sly)
LOC103626462 (zma)
LOC103864240 (bra)
|
Subcellular
localization
wolf |
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_001332642.1)
|
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_001332643.1)
|
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_001332644.1)
|
|
plas 7,
E.R. 1,
nucl 1,
vacu 1,
cyto_E.R. 1
|
(predict for NP_198898.2)
|
|
Subcellular
localization
TargetP |
|
other 5
|
(predict for NP_001332642.1)
|
|
other 5
|
(predict for NP_001332643.1)
|
|
other 5
|
(predict for NP_001332644.1)
|
|
other 5
|
(predict for NP_198898.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04120 |
Ubiquitin mediated proteolysis |
2 |
 |
| ath03030 |
DNA replication |
2 |
 |
| ath03410 |
Base excision repair |
2 |
 |
| ath03420 |
Nucleotide excision repair |
2 |
 |
| ath03430 |
Mismatch repair |
2 |
 |
Genes directly connected with ATR on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.5 |
AT2G04235 |
hypothetical protein |
[detail] |
814961 |
| 6.5 |
AT4G36080 |
phosphotransferases/inositol or phosphatidylinositol kinase |
[detail] |
829764 |
| 5.8 |
AT5G63960 |
DNA polymerase delta subunit 1 |
[detail] |
836517 |
| 4.6 |
AT4G02660 |
Beige/BEACH and WD40 domain-containing protein |
[detail] |
828210 |
| 3.8 |
XIC |
Myosin family protein with Dil domain-containing protein |
[detail] |
837394 |
|
Coexpressed
gene list |
[Coexpressed gene list for ATR]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
249328_at
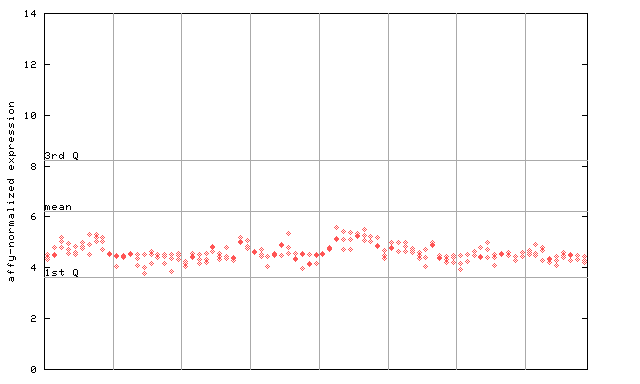
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
249328_at
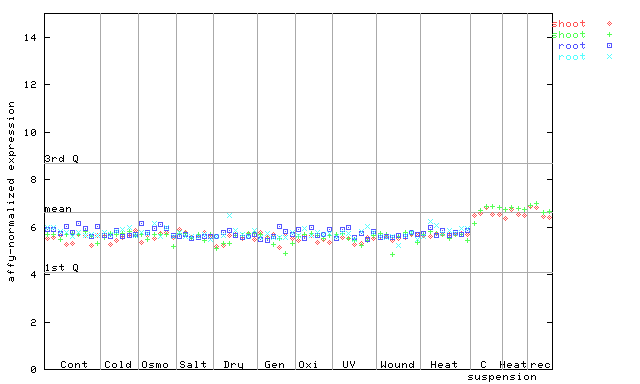
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
249328_at
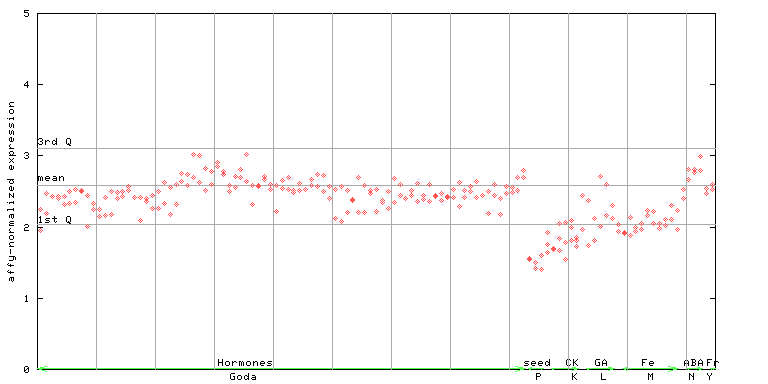
X axis is samples (xls file), and Y axis is log-expression.
|

















