| functional annotation |
| Function |
Leucine-rich receptor-like protein kinase family protein |


|
| GO BP |
|
GO:0010480 [list] [network] microsporocyte differentiation
|
(2 genes)
|
IGI
|
 |
|
GO:0009934 [list] [network] regulation of meristem structural organization
|
(21 genes)
|
IGI
|
 |
|
GO:0010075 [list] [network] regulation of meristem growth
|
(54 genes)
|
IGI
|
 |
|
GO:0048653 [list] [network] anther development
|
(75 genes)
|
IGI
|
 |
|
GO:0048437 [list] [network] floral organ development
|
(242 genes)
|
IGI
|
 |
|
GO:0048229 [list] [network] gametophyte development
|
(446 genes)
|
IGI
|
 |
|
GO:0030154 [list] [network] cell differentiation
|
(695 genes)
|
IEA
|
 |
|
GO:0006468 [list] [network] protein phosphorylation
|
(921 genes)
|
IEA
|
 |
|
| GO CC |
|
GO:0005886 [list] [network] plasma membrane
|
(3771 genes)
|
IBA
IDA
ISM
|
 |
|
GO:0016021 [list] [network] integral component of membrane
|
(4803 genes)
|
IEA
|
 |
|
| GO MF |
|
GO:0033612 [list] [network] receptor serine/threonine kinase binding
|
(38 genes)
|
IPI
|
 |
|
GO:0043621 [list] [network] protein self-association
|
(96 genes)
|
IDA
|
 |
|
GO:0042802 [list] [network] identical protein binding
|
(288 genes)
|
IPI
|
 |
|
GO:0004674 [list] [network] protein serine/threonine kinase activity
|
(801 genes)
|
IEA
|
 |
|
GO:0016301 [list] [network] kinase activity
|
(1362 genes)
|
ISS
|
 |
|
GO:0005524 [list] [network] ATP binding
|
(2003 genes)
|
IEA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001190624.1
NP_201371.1
|
| BLAST |
NP_001190624.1
NP_201371.1
|
| Orthologous |
[Ortholog page]
RLK1 (gma)
RLK3 (gma)
RLK2 (gma)
BAM2 (ath)
BAM3 (ath)
LOC4332141 (osa)
LOC4334273 (osa)
LOC4342345 (osa)
LOC7465379 (ppo)
LOC7465545 (ppo)
LOC11408427 (mtr)
LOC11424811 (mtr)
LOC11431479 (mtr)
LOC25490539 (mtr)
LOC100258232 (vvi)
LOC100279272 (zma)
LOC100383797 (zma)
LOC100499646 (gma)
LOC100777902 (gma)
LOC100803075 (gma)
LOC100815103 (gma)
LOC100816158 (gma)
LOC100855393 (vvi)
LOC101248733 (sly)
LOC101248913 (sly)
LOC101264597 (sly)
LOC103644083 (zma)
LOC103834155 (bra)
LOC103858059 (bra)
LOC103860625 (bra)
LOC103861192 (bra)
LOC103874010 (bra)
LOC109939609 (zma)
|
Subcellular
localization
wolf |
|
plas 7,
E.R. 1,
chlo 1,
pero 1,
E.R._vacu 1,
cyto_E.R. 1
|
(predict for NP_001190624.1)
|
|
plas 7,
E.R. 1,
chlo 1,
pero 1,
E.R._vacu 1,
cyto_E.R. 1
|
(predict for NP_201371.1)
|
|
Subcellular
localization
TargetP |
|
scret 2
|
(predict for NP_001190624.1)
|
|
scret 2
|
(predict for NP_201371.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with BAM1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.6 |
AT3G56370 |
Leucine-rich repeat protein kinase family protein |
[detail] |
824804 |
| 6.3 |
BAM2 |
Leucine-rich receptor-like protein kinase family protein |
[detail] |
824129 |
| 6.3 |
ALE2 |
Protein kinase superfamily protein |
[detail] |
816549 |
| 6.1 |
SLP2 |
subtilisin-like serine protease 2 |
[detail] |
829650 |
| 6.0 |
HSL1 |
HAESA-like 1 |
[detail] |
839742 |
| 5.8 |
AT5G66770 |
GRAS family transcription factor |
[detail] |
836810 |
| 5.7 |
AT5G62890 |
Xanthine/uracil permease family protein |
[detail] |
836409 |
| 5.3 |
AT2G05920 |
Subtilase family protein |
[detail] |
815145 |
| 5.2 |
AT5G56040 |
Leucine-rich receptor-like protein kinase family protein |
[detail] |
835702 |
| 3.8 |
MAKR2 |
membrane-associated kinase regulator |
[detail] |
842712 |
|
Coexpressed
gene list |
[Coexpressed gene list for BAM1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
247153_at
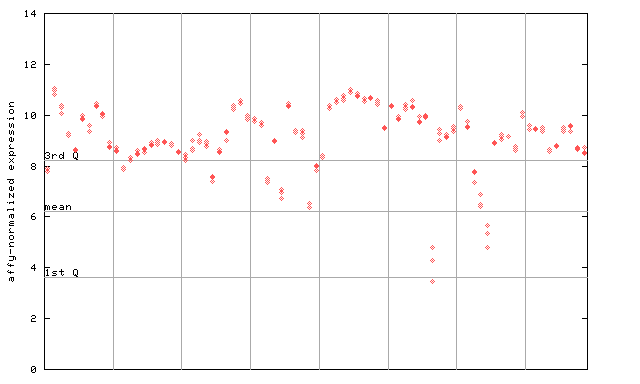
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
247153_at
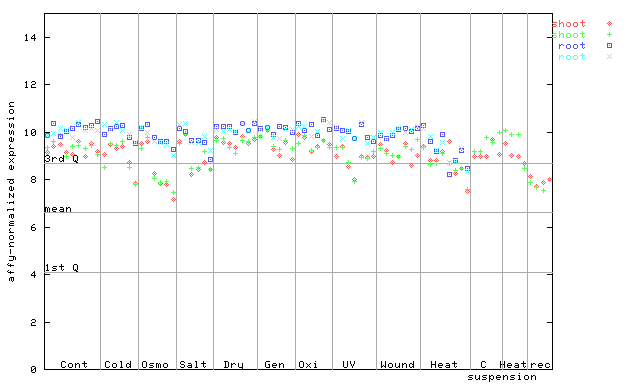
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
247153_at
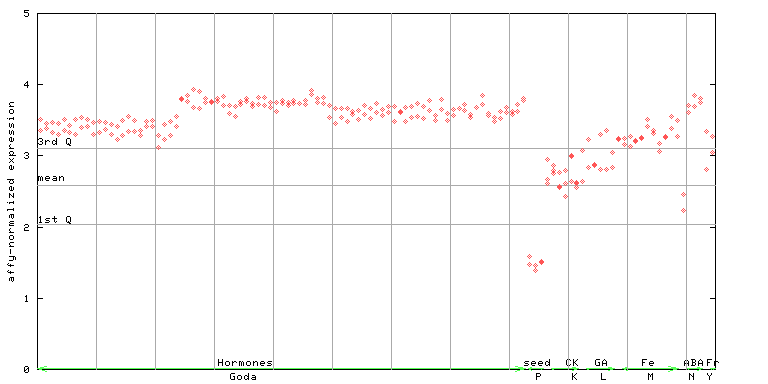
X axis is samples (xls file), and Y axis is log-expression.
|










