[←][→] ath
| functional annotation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | cell wall-associated kinase |


|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_564137.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_564137.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] AT3G25490 (ath) WAK4 (ath) WAK5 (ath) WAK3 (ath) AT1G21245 (ath) WAK2 (ath) AT1G22720 (ath) LOC4325722 (osa) LOC4328047 (osa) LOC4330026 (osa) LOC4331078 (osa) LOC4331082 (osa) LOC4331083 (osa) LOC4331100 (osa) LOC4335352 (osa) LOC4335580 (osa) LOC4336862 (osa) LOC4336863 (osa) LOC4340086 (osa) LOC4347337 (osa) LOC4347339 (osa) LOC4347341 (osa) LOC4352786 (osa) LOC4352788 (osa) LOC4352790 (osa) LOC9266348 (osa) LOC9268903 (osa) LOC9270668 (osa) LOC11423592 (mtr) LOC25481907 (mtr) LOC25481909 (mtr) LOC25481910 (mtr) LOC25481911 (mtr) LOC25482293 (mtr) LOC25482298 (mtr) LOC25482301 (mtr) LOC25482303 (mtr) LOC25482305 (mtr) LOC25482306 (mtr) LOC25489724 (mtr) LOC25489725 (mtr) LOC25489726 (mtr) LOC25489728 (mtr) LOC100191438 (zma) LOC100247712 (vvi) LOC100252247 (vvi) LOC100252663 (vvi) LOC100259679 (vvi) LOC100280308 (zma) LOC100280667 (zma) LOC100281741 (zma) LOC100285958 (zma) LOC100384815 (zma) LOC100775699 (gma) LOC100776778 (gma) LOC100778914 (gma) LOC100779983 (gma) LOC100781051 (gma) LOC100784078 (gma) LOC100785674 (gma) LOC100797492 (gma) LOC100798027 (gma) LOC100819535 (gma) LOC100852995 (vvi) LOC101247359 (sly) LOC101258182 (sly) LOC101262650 (sly) LOC101262780 (sly) LOC101263242 (sly) LOC103627429 (zma) LOC103646213 (zma) LOC103646216 (zma) LOC103648521 (zma) LOC103649276 (zma) LOC103829384 (bra) LOC103829385 (bra) LOC103835750 (bra) LOC103835752 (bra) LOC103835753 (bra) LOC103872913 (bra) LOC103872914 (bra) LOC103872915 (bra) LOC104649046 (sly) LOC106795446 (gma) LOC106795885 (gma) LOC107275972 (osa) LOC107276036 (osa) LOC107276382 (osa) LOC107276480 (osa) LOC109941939 (zma) LOC113001466 (gma) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for WAK1] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
259561_at
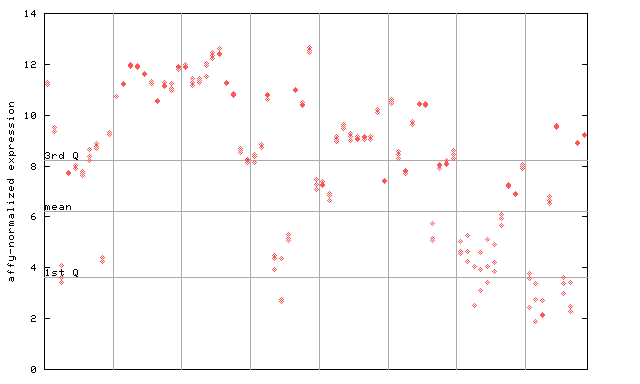
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
259561_at
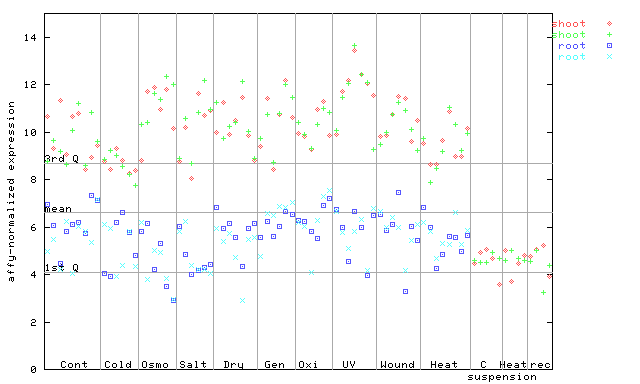
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
259561_at
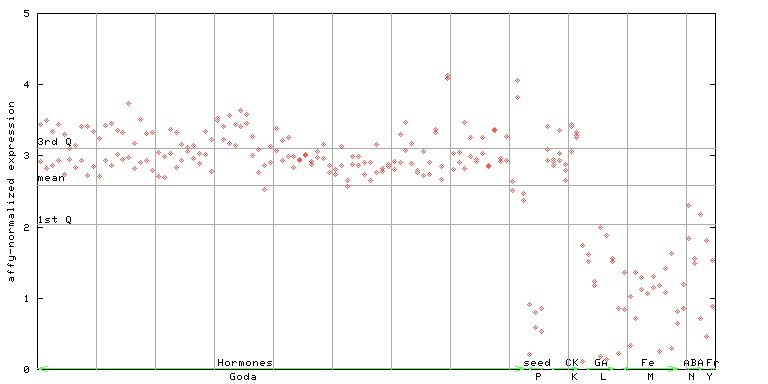
X axis is samples (xls file), and Y axis is log-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 838721 |


|
| Refseq ID (protein) | NP_564137.1 |  |
The preparation time of this page was 0.2 [sec].


