[←][→] ath
| functional annotation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | Leucine-rich repeat protein kinase family protein |


|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_001319202.1 NP_001322768.1 NP_001322769.1 NP_001322770.1 NP_175601.2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_001319202.1 NP_001322768.1 NP_001322769.1 NP_001322770.1 NP_175601.2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] AT2G04300 (ath) AT2G14440 (ath) AT2G14510 (ath) FRK1 (ath) AT2G19210 (ath) AT2G19230 (ath) AT2G28960 (ath) AT2G28970 (ath) AT2G28990 (ath) AT2G29000 (ath) AT3G21340 (ath) AT3G46350 (ath) AT3G46370 (ath) RHS16 (ath) AT4G29450 (ath) AT4G29990 (ath) AT5G16900 (ath) AT5G59650 (ath) AT5G59670 (ath) AT5G59680 (ath) AT1G05700 (ath) AT1G07550 (ath) AT1G07560 (ath) AT1G49100 (ath) AT1G51790 (ath) IOS1 (ath) AT1G51805 (ath) AT1G51810 (ath) AT1G51820 (ath) AT1G51830 (ath) AT1G51840 (ath) AT1G51850 (ath) AT1G51860 (ath) AT1G51870 (ath) RHS6 (ath) AT1G51910 (ath) LOC4328315 (osa) LOC4339370 (osa) LOC4339371 (osa) LOC4339374 (osa) LOC4339376 (osa) LOC4339378 (osa) LOC4346817 (osa) LOC9270191 (osa) LOC9272354 (osa) LOC11421236 (mtr) LOC11422007 (mtr) LOC11425516 (mtr) LOC11425517 (mtr) LOC11429446 (mtr) LOC11433438 (mtr) LOC11434037 (mtr) LOC25497008 (mtr) LOC25500355 (mtr) LOC25500358 (mtr) LOC25500359 (mtr) LOC25500361 (mtr) LOC25500364 (mtr) LOC25500366 (mtr) LOC25500367 (mtr) LOC25500376 (mtr) LOC25500764 (mtr) LOC25500765 (mtr) LOC25502465 (mtr) SIK2 (gma) LOC100242932 (vvi) LOC100244634 (vvi) LOC100246311 (vvi) LOC100246405 (vvi) LOC100249701 (vvi) LOC100251452 (vvi) LOC100260016 (vvi) LOC100261686 (vvi) LOC100266874 (vvi) LOC100266960 (vvi) LOC100275543 (zma) LOC100775741 (gma) LOC100776284 (gma) LOC100776818 (gma) LOC100778537 (gma) LOC100779102 (gma) LOC100779474 (gma) LOC100779594 (gma) LOC100779617 (gma) LOC100781084 (gma) LOC100781785 (gma) LOC100785345 (gma) LOC100791717 (gma) LOC100792239 (gma) LOC100793137 (gma) LOC100794504 (gma) LOC100795183 (gma) LOC100818475 (gma) LOC101257947 (sly) LOC101262429 (sly) LOC101266705 (sly) LOC102660013 (gma) LOC102662730 (gma) LOC102663143 (gma) LOC103626627 (zma) LOC103634037 (zma) LOC103829493 (bra) LOC103829497 (bra) LOC103836435 (bra) LOC103843445 (bra) LOC103843938 (bra) LOC103844100 (bra) LOC103845385 (bra) LOC103845386 (bra) LOC103850089 (bra) LOC103851561 (bra) LOC103851563 (bra) LOC103851564 (bra) LOC103851565 (bra) LOC103853036 (bra) LOC103853548 (bra) LOC103854870 (bra) LOC103856616 (bra) LOC103856900 (bra) LOC103859903 (bra) LOC103861877 (bra) LOC103864933 (bra) LOC103864934 (bra) LOC103868533 (bra) LOC103868536 (bra) LOC103868540 (bra) LOC103868542 (bra) LOC103868543 (bra) LOC103868545 (bra) LOC103868632 (bra) LOC103869180 (bra) LOC103871250 (bra) LOC103871251 (bra) LOC103871253 (bra) LOC103871255 (bra) LOC104880223 (vvi) LOC104880225 (vvi) LOC106794186 (gma) LOC106799536 (gma) LOC107275782 (osa) LOC107276230 (osa) LOC107278846 (osa) LOC108870913 (bra) LOC112416101 (mtr) LOC112417465 (mtr) LOC112417487 (mtr) LOC113002309 (gma) LOC113002392 (gma) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for AT1G51890] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
246368_at
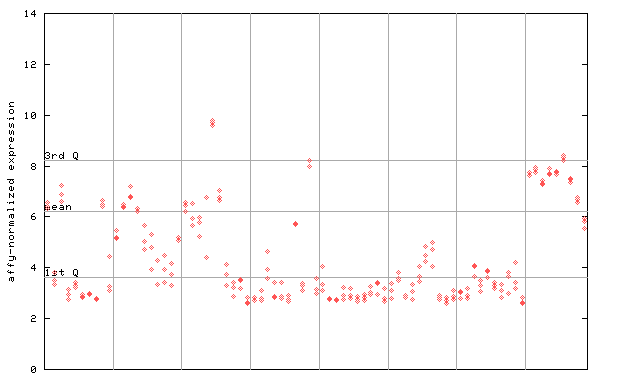
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
246368_at
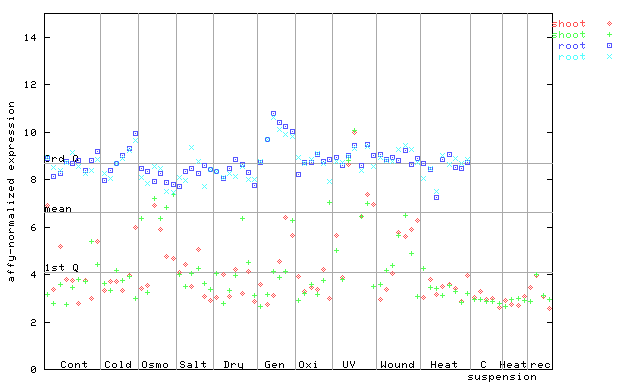
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
246368_at
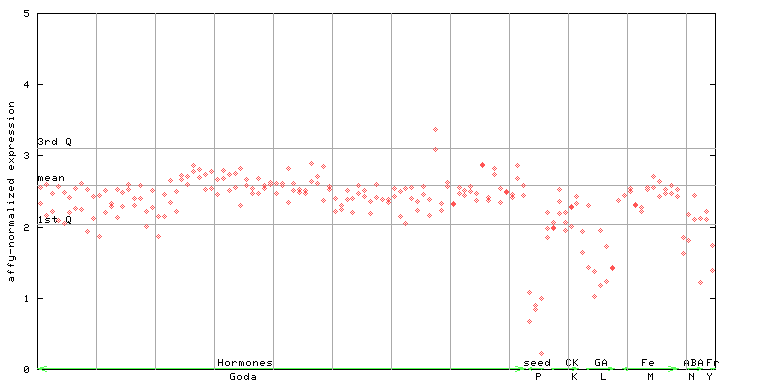
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 841616 |


|
| Refseq ID (protein) | NP_001319202.1 |  |
| NP_001322768.1 |  |
|
| NP_001322769.1 |  |
|
| NP_001322770.1 |  |
|
| NP_175601.2 |  |
|
The preparation time of this page was 0.2 [sec].


