| functional annotation |
| Function |
exoribonuclease 4 |


|
| GO BP |
|
GO:0009961 [list] [network] response to 1-aminocyclopropane-1-carboxylic acid
|
(5 genes)
|
IMP
|
 |
|
GO:0010587 [list] [network] miRNA catabolic process
|
(6 genes)
|
IMP
|
 |
|
GO:0031087 [list] [network] deadenylation-independent decapping of nuclear-transcribed mRNA
|
(6 genes)
|
IMP
|
 |
|
GO:0061014 [list] [network] positive regulation of mRNA catabolic process
|
(6 genes)
|
IGI
|
 |
|
GO:0070370 [list] [network] cellular heat acclimation
|
(10 genes)
|
IEP
IMP
|
 |
|
GO:0000291 [list] [network] nuclear-transcribed mRNA catabolic process, exonucleolytic
|
(24 genes)
|
IMP
|
 |
|
GO:0010286 [list] [network] heat acclimation
|
(53 genes)
|
IMP
|
 |
|
GO:0006402 [list] [network] mRNA catabolic process
|
(80 genes)
|
IMP
|
 |
|
GO:0009630 [list] [network] gravitropism
|
(90 genes)
|
IMP
|
 |
|
GO:0010087 [list] [network] phloem or xylem histogenesis
|
(102 genes)
|
IMP
|
 |
|
GO:0009873 [list] [network] ethylene-activated signaling pathway
|
(191 genes)
|
IGI
IMP
|
 |
|
GO:0040029 [list] [network] regulation of gene expression, epigenetic
|
(210 genes)
|
IMP
|
 |
|
GO:0009826 [list] [network] unidimensional cell growth
|
(279 genes)
|
IMP
|
 |
|
GO:0009723 [list] [network] response to ethylene
|
(297 genes)
|
IMP
|
 |
|
GO:0051301 [list] [network] cell division
|
(352 genes)
|
IMP
|
 |
|
GO:0006397 [list] [network] mRNA processing
|
(376 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
ath04016 [list] [network] MAPK signaling pathway - plant (134 genes) |
 |
| Protein |
NP_001321327.1
NP_175851.1
|
| BLAST |
NP_001321327.1
NP_175851.1
|
| Orthologous |
[Ortholog page]
LOC4334408 (osa)
LOC11429796 (mtr)
XRN4 (sly)
LOC100253751 (vvi)
LOC100384297 (zma)
LOC100789497 (gma)
LOC100793385 (gma)
LOC100817715 (gma)
LOC103838732 (bra)
LOC112421269 (mtr)
|
Subcellular
localization
wolf |
|
cyto 5,
nucl 4,
chlo 1,
plas 1
|
(predict for NP_001321327.1)
|
|
cyto 9
|
(predict for NP_175851.1)
|
|
Subcellular
localization
TargetP |
|
other 8
|
(predict for NP_001321327.1)
|
|
other 7,
mito 4
|
(predict for NP_175851.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with XRN4 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 7.9 |
UBP13 |
ubiquitin-specific protease 13 |
[detail] |
820364 |
| 7.4 |
AT5G07940 |
dentin sialophosphoprotein-like protein |
[detail] |
830688 |
| 7.2 |
AT1G02080 |
transcription regulator |
[detail] |
839244 |
| 6.8 |
AT5G42220 |
Ubiquitin-like superfamily protein |
[detail] |
834227 |
| 5.9 |
ZIGA4 |
ARF GAP-like zinc finger-containing protein ZIGA4 |
[detail] |
837390 |
|
Coexpressed
gene list |
[Coexpressed gene list for XRN4]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
262961_at
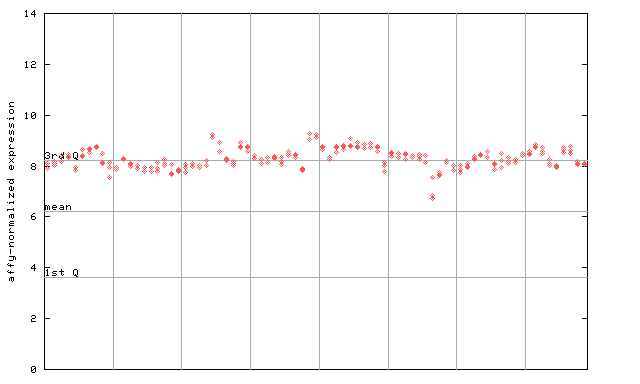
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
262961_at
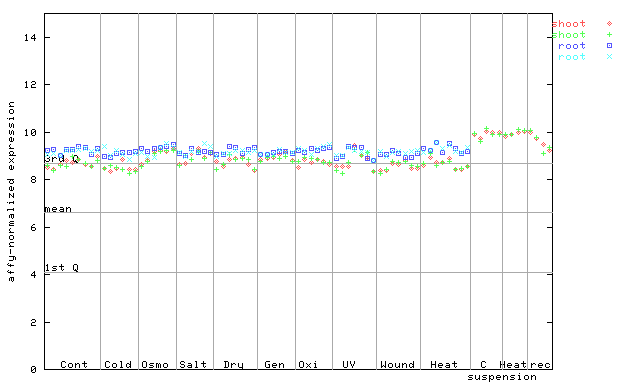
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
262961_at
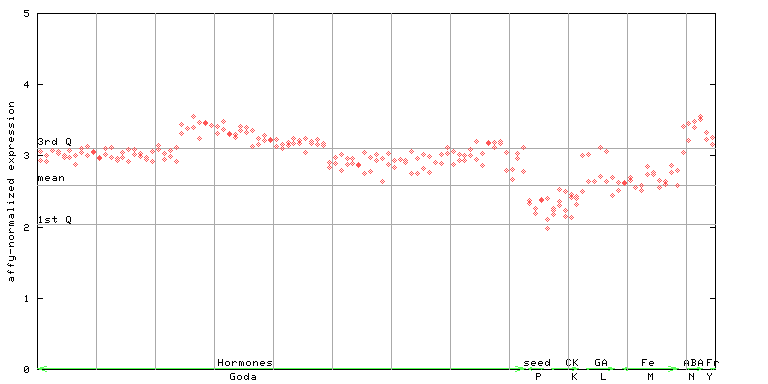
X axis is samples (xls file), and Y axis is log-expression.
|











