| functional annotation |
| Function |
Signal transduction histidine kinase, hybrid-type, ethylene sensor |


|
| GO BP |
|
GO:0009727 [list] [network] detection of ethylene stimulus
|
(3 genes)
|
IMP
|
 |
|
GO:0009871 [list] [network] jasmonic acid and ethylene-dependent systemic resistance, ethylene mediated signaling pathway
|
(4 genes)
|
TAS
|
 |
|
GO:0050665 [list] [network] hydrogen peroxide biosynthetic process
|
(9 genes)
|
IMP
|
 |
|
GO:0052544 [list] [network] defense response by callose deposition in cell wall
|
(16 genes)
|
IMP
|
 |
|
GO:0010105 [list] [network] negative regulation of ethylene-activated signaling pathway
|
(17 genes)
|
TAS
|
 |
|
GO:0009625 [list] [network] response to insect
|
(32 genes)
|
IMP
|
 |
|
GO:0009690 [list] [network] cytokinin metabolic process
|
(33 genes)
|
IMP
|
 |
|
GO:0002237 [list] [network] response to molecule of bacterial origin
|
(34 genes)
|
IMP
|
 |
|
GO:0010182 [list] [network] sugar mediated signaling pathway
|
(37 genes)
|
TAS
|
 |
|
GO:1900140 [list] [network] regulation of seedling development
|
(87 genes)
|
IMP
|
 |
|
GO:0010119 [list] [network] regulation of stomatal movement
|
(88 genes)
|
IMP
|
 |
|
GO:0010087 [list] [network] phloem or xylem histogenesis
|
(102 genes)
|
IMP
|
 |
|
GO:0009739 [list] [network] response to gibberellin
|
(154 genes)
|
IMP
|
 |
|
GO:0009873 [list] [network] ethylene-activated signaling pathway
|
(191 genes)
|
IEA
|
 |
|
GO:0009408 [list] [network] response to heat
|
(229 genes)
|
IMP
|
 |
|
GO:0009723 [list] [network] response to ethylene
|
(297 genes)
|
IMP
|
 |
|
GO:0051301 [list] [network] cell division
|
(352 genes)
|
IMP
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(399 genes)
|
IMP
|
 |
|
GO:0009733 [list] [network] response to auxin
|
(407 genes)
|
IMP
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(485 genes)
|
IEP
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(574 genes)
|
IMP
|
 |
|
GO:0006952 [list] [network] defense response
|
(1420 genes)
|
TAS
|
 |
|
| GO CC |
|
GO:0005789 [list] [network] endoplasmic reticulum membrane
|
(466 genes)
|
IDA
IEA
|
 |
|
GO:0005783 [list] [network] endoplasmic reticulum
|
(877 genes)
|
IBA
IDA
|
 |
|
GO:0016021 [list] [network] integral component of membrane
|
(4803 genes)
|
IEA
|
 |
|
| GO MF |
|
| KEGG |
ath04016 [list] [network] MAPK signaling pathway - plant (134 genes) |
 |
| ath04075 [list] [network] Plant hormone signal transduction (273 genes) |
 |
| Protein |
NP_176808.3
|
| BLAST |
NP_176808.3
|
| Orthologous |
[Ortholog page]
NR (sly)
ETR1 (sly)
ETR2 (sly)
ERS1 (ath)
LOC4333832 (osa)
LOC4337850 (osa)
LOC7478853 (ppo)
LOC11431567 (mtr)
LOC11434695 (mtr)
LOC100126971 (zma)
LOC100217201 (zma)
ETR1 (vvi)
LOC100241730 (vvi)
LOC100794417 (gma)
LOC100795340 (gma)
LOC100795799 (gma)
LOC100802569 (gma)
LOC103630068 (zma)
ETR1 (bra)
LOC103866369 (bra)
|
Subcellular
localization
wolf |
|
cyto 6,
plas 2,
nucl 1,
E.R. 1,
cysk 1,
cysk_nucl 1
|
(predict for NP_176808.3)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_176808.3)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with ETR1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.4 |
AT3G05545 |
RING/U-box superfamily protein |
[detail] |
819720 |
| 6.0 |
UBP16 |
ubiquitin-specific protease 16 |
[detail] |
828558 |
| 6.0 |
SUO |
protein SUO |
[detail] |
823960 |
| 5.4 |
AT5G11850 |
Protein kinase superfamily protein |
[detail] |
831058 |
|
Coexpressed
gene list |
[Coexpressed gene list for ETR1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
260133_at
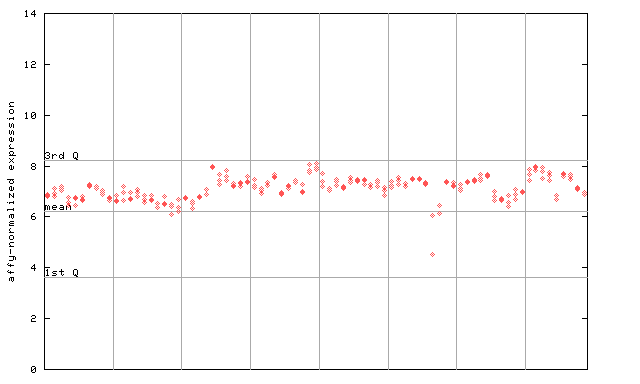
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
260133_at
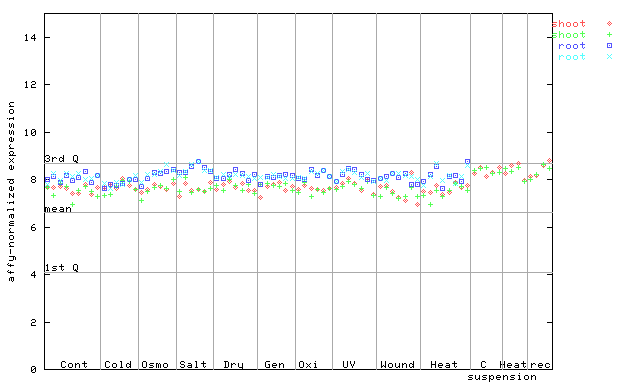
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
260133_at
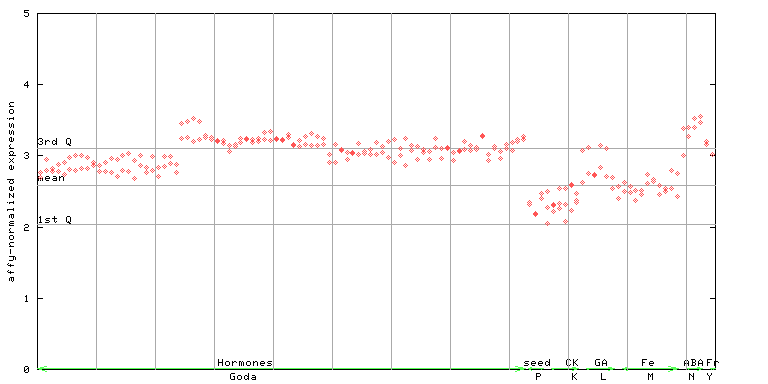
X axis is samples (xls file), and Y axis is log-expression.
|











