| functional annotation |
| Function |
vascular related NAC-domain protein 7 |


|
| GO BP |
|
GO:0090059 [list] [network] protoxylem development
|
(3 genes)
|
IMP
|
 |
|
GO:0048759 [list] [network] xylem vessel member cell differentiation
|
(11 genes)
|
IDA
IMP
|
 |
|
GO:0010089 [list] [network] xylem development
|
(42 genes)
|
IMP
TAS
|
 |
|
GO:0045491 [list] [network] xylan metabolic process
|
(52 genes)
|
IMP
|
 |
|
GO:0009735 [list] [network] response to cytokinin
|
(102 genes)
|
IEP
|
 |
|
GO:0009741 [list] [network] response to brassinosteroid
|
(109 genes)
|
IEP
|
 |
|
GO:0071365 [list] [network] cellular response to auxin stimulus
|
(215 genes)
|
IDA
|
 |
|
GO:0009733 [list] [network] response to auxin
|
(407 genes)
|
IEP
|
 |
|
GO:0045893 [list] [network] positive regulation of transcription, DNA-templated
|
(495 genes)
|
IDA
|
 |
|
GO:0050832 [list] [network] defense response to fungus
|
(517 genes)
|
IMP
|
 |
|
GO:0071555 [list] [network] cell wall organization
|
(538 genes)
|
IEA
|
 |
|
GO:0010628 [list] [network] positive regulation of gene expression
|
(561 genes)
|
IDA
IMP
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(574 genes)
|
IEP
|
 |
|
GO:0009620 [list] [network] response to fungus
|
(595 genes)
|
IDA
|
 |
|
GO:0006355 [list] [network] regulation of transcription, DNA-templated
|
(1984 genes)
|
TAS
|
 |
|
GO:0007275 [list] [network] multicellular organism development
|
(2714 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0046982 [list] [network] protein heterodimerization activity
|
(122 genes)
|
IDA
|
 |
|
GO:0042803 [list] [network] protein homodimerization activity
|
(158 genes)
|
IDA
|
 |
|
GO:0042802 [list] [network] identical protein binding
|
(288 genes)
|
IPI
|
 |
|
GO:0043565 [list] [network] sequence-specific DNA binding
|
(796 genes)
|
IDA
|
 |
|
GO:0044212 [list] [network] transcription regulatory region DNA binding
|
(871 genes)
|
IPI
|
 |
|
GO:0003700 [list] [network] DNA-binding transcription factor activity
|
(1599 genes)
|
IDA
IMP
ISS
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001323397.1
NP_177338.1
|
| BLAST |
NP_001323397.1
NP_177338.1
|
| Orthologous |
[Ortholog page]
LOC4344435 (osa)
LOC25484969 (mtr)
LOC100192643 (zma)
LOC100265732 (vvi)
LOC100281942 (zma)
LOC100811057 (gma)
GMNAC184 (gma)
LOC101245609 (sly)
LOC101252786 (sly)
LOC103653304 (zma)
LOC103831706 (bra)
LOC103852738 (bra)
|
Subcellular
localization
wolf |
|
nucl 8,
cyto_nucl 4,
chlo 1,
plas 1
|
(predict for NP_001323397.1)
|
|
nucl 8,
cyto_nucl 4,
chlo 1,
plas 1
|
(predict for NP_177338.1)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_001323397.1)
|
|
other 9
|
(predict for NP_177338.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
|
Coexpressed
gene list |
[Coexpressed gene list for VND7]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
260173_at
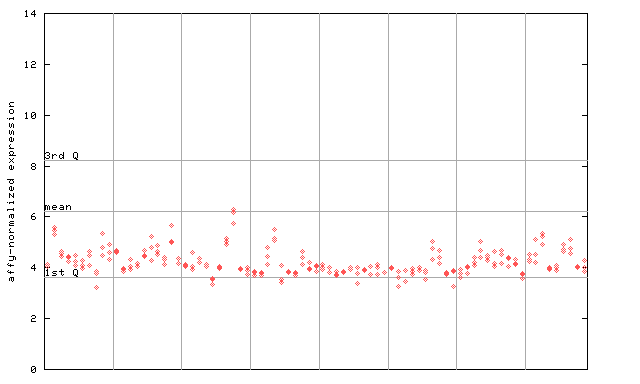
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
260173_at
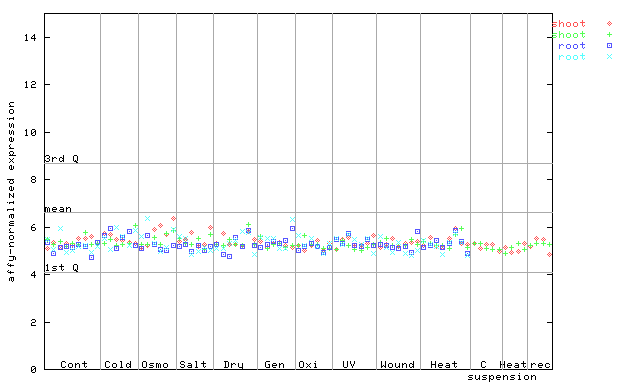
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
260173_at
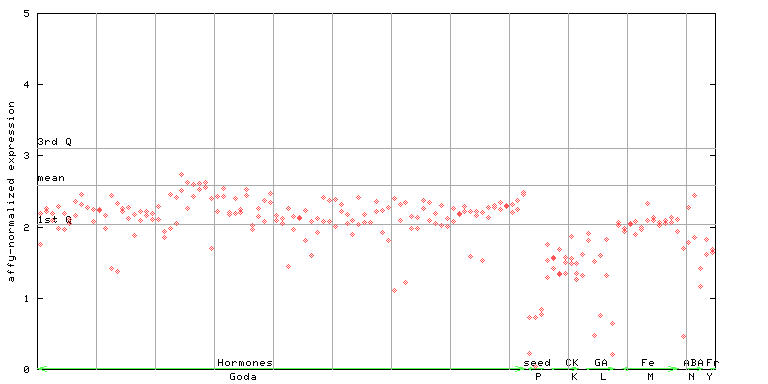
X axis is samples (xls file), and Y axis is log-expression.
|










