| functional annotation |
| Function |
nuclear pore anchor |


|
| GO BP |
|
GO:0033234 [list] [network] negative regulation of protein sumoylation
|
(1 genes)
|
IMP
|
 |
|
GO:0016973 [list] [network] poly(A)+ mRNA export from nucleus
|
(10 genes)
|
IMP
|
 |
|
GO:0007094 [list] [network] mitotic spindle assembly checkpoint
|
(11 genes)
|
IBA
|
 |
|
GO:0006606 [list] [network] protein import into nucleus
|
(47 genes)
|
IEA
|
 |
|
GO:0009910 [list] [network] negative regulation of flower development
|
(50 genes)
|
IMP
|
 |
|
GO:0060968 [list] [network] regulation of gene silencing
|
(60 genes)
|
IMP
|
 |
|
GO:0048443 [list] [network] stamen development
|
(101 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0017056 [list] [network] structural constituent of nuclear pore
|
(21 genes)
|
IBA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
ath03013 [list] [network] RNA transport (171 genes) |
 |
| Protein |
NP_001185435.1
NP_001185436.1
NP_178048.2
|
| BLAST |
NP_001185435.1
NP_001185436.1
NP_178048.2
|
| Orthologous |
[Ortholog page]
LOC4330687 (osa)
LOC7460662 (ppo)
LOC11418624 (mtr)
LOC100193455 (zma)
LOC100251875 (vvi)
LOC100788022 (gma)
LOC100811882 (gma)
LOC101248882 (sly)
LOC103830490 (bra)
LOC103832403 (bra)
|
Subcellular
localization
wolf |
|
nucl 6,
cyto 2,
cysk 1,
golg 1
|
(predict for NP_001185435.1)
|
|
nucl 6,
cyto 2,
cysk 1,
golg 1
|
(predict for NP_001185436.1)
|
|
nucl 6,
cyto 2,
cysk 1,
golg 1
|
(predict for NP_178048.2)
|
|
Subcellular
localization
TargetP |
|
other 8
|
(predict for NP_001185435.1)
|
|
other 8
|
(predict for NP_001185436.1)
|
|
other 8
|
(predict for NP_178048.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03013 |
RNA transport |
2 |
 |
Genes directly connected with NUA on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 14.1 |
THO2 |
THO2 |
[detail] |
839081 |
| 13.0 |
AT3G47910 |
Ubiquitin carboxyl-terminal hydrolase-related protein |
[detail] |
823946 |
| 11.5 |
CHR5 |
chromatin remodeling 5 |
[detail] |
815823 |
| 11.4 |
emb1579 |
ATP/GTP-binding protein family |
[detail] |
814844 |
| 10.3 |
MOR1 |
ARM repeat superfamily protein |
[detail] |
818131 |
| 10.1 |
RAD50 |
DNA repair-recombination protein (RAD50) |
[detail] |
817756 |
| 9.9 |
ATRX |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
837382 |
| 9.2 |
LINC2 |
nuclear matrix constituent protein-like protein |
[detail] |
837882 |
| 7.9 |
CIP4.1 |
COP1-interacting protein 4.1 |
[detail] |
827980 |
| 7.9 |
AT3G19740 |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
821511 |
| 7.7 |
AT1G16800 |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
838251 |
| 6.7 |
SNL6 |
paired amphipathic helix Sin3-like protein |
[detail] |
837584 |
|
Coexpressed
gene list |
[Coexpressed gene list for NUA]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
264126_at
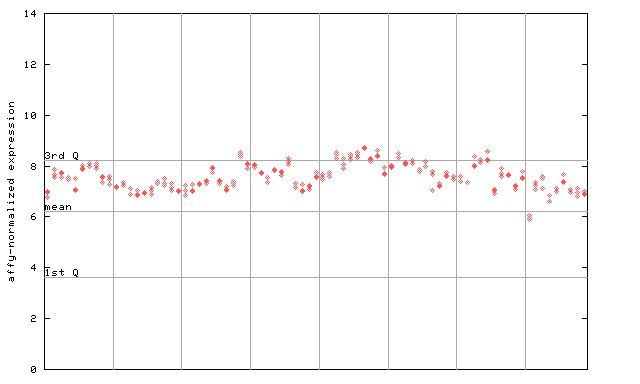
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
264126_at
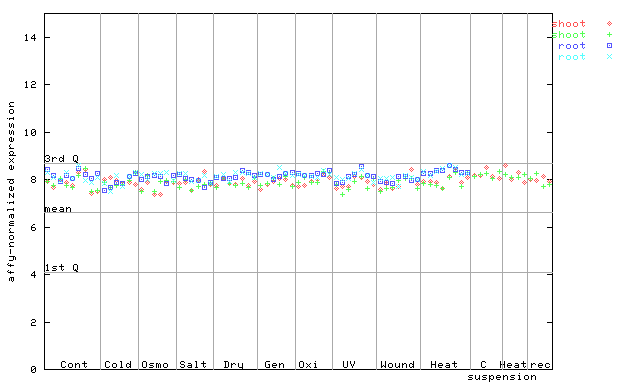
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
264126_at
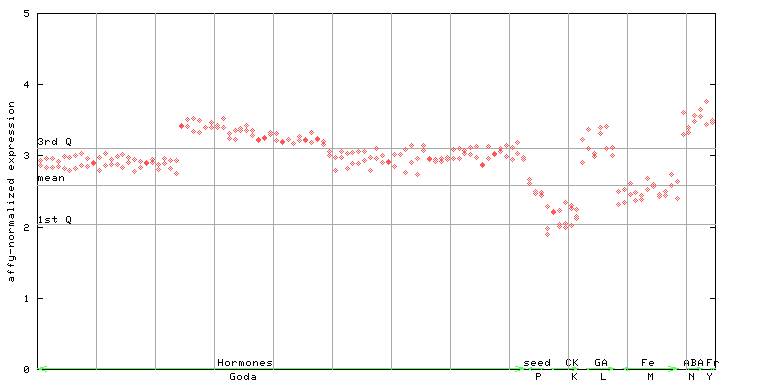
X axis is samples (xls file), and Y axis is log-expression.
|













