| functional annotation |
| Function |
RNAligase |


|
| GO BP |
|
GO:0010069 [list] [network] zygote asymmetric cytokinesis in embryo sac
|
(6 genes)
|
IMP
|
 |
|
GO:0006388 [list] [network] tRNA splicing, via endonucleolytic cleavage and ligation
|
(10 genes)
|
IDA
IMP
TAS
|
 |
|
GO:0030968 [list] [network] endoplasmic reticulum unfolded protein response
|
(29 genes)
|
IDA
|
 |
|
GO:0010928 [list] [network] regulation of auxin mediated signaling pathway
|
(30 genes)
|
IMP
|
 |
|
GO:0009734 [list] [network] auxin-activated signaling pathway
|
(196 genes)
|
IEA
|
 |
|
GO:0006412 [list] [network] translation
|
(1149 genes)
|
IEA
|
 |
|
GO:0016310 [list] [network] phosphorylation
|
(1279 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0003972 [list] [network] RNA ligase (ATP) activity
|
(1 genes)
|
IDA
IGI
|
 |
|
GO:0004113 [list] [network] 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity
|
(3 genes)
|
IGI
|
 |
|
GO:0051731 [list] [network] polynucleotide 5'-hydroxyl-kinase activity
|
(5 genes)
|
IGI
|
 |
|
GO:0004519 [list] [network] endonuclease activity
|
(145 genes)
|
IEA
|
 |
|
GO:0003729 [list] [network] mRNA binding
|
(1026 genes)
|
IDA
|
 |
|
GO:0016301 [list] [network] kinase activity
|
(1362 genes)
|
IEA
|
 |
|
GO:0005524 [list] [network] ATP binding
|
(2003 genes)
|
IEA
|
 |
|
GO:0046872 [list] [network] metal ion binding
|
(3180 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001318950.1
NP_172269.2
|
| BLAST |
NP_001318950.1
NP_172269.2
|
| Orthologous |
[Ortholog page]
LOC4342624 (osa)
LOC25492457 (mtr)
LOC100258617 (vvi)
LOC100815563 (gma)
LOC101247886 (sly)
LOC103632082 (zma)
LOC103843428 (bra)
LOC103871536 (bra)
|
Subcellular
localization
wolf |
|
chlo 10
|
(predict for NP_001318950.1)
|
|
nucl 8,
chlo 1,
cyto 1,
plas 1,
cyto_plas 1
|
(predict for NP_172269.2)
|
|
Subcellular
localization
TargetP |
|
chlo 9
|
(predict for NP_001318950.1)
|
|
chlo 6,
other 6
|
(predict for NP_172269.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03013 |
RNA transport |
3 |
 |
| ath03040 |
Spliceosome |
2 |
 |
Genes directly connected with RNL on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 7.3 |
AT3G19740 |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
821511 |
| 6.8 |
AT5G11350 |
DNAse I-like superfamily protein |
[detail] |
831006 |
| 6.1 |
AT1G26170 |
ARM repeat superfamily protein |
[detail] |
839158 |
|
Coexpressed
gene list |
[Coexpressed gene list for RNL]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
260677_at
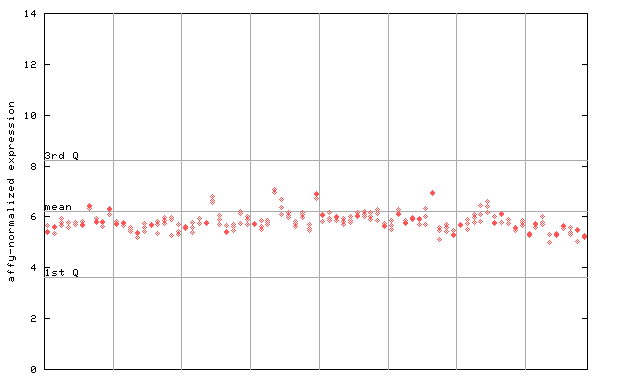
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
260677_at
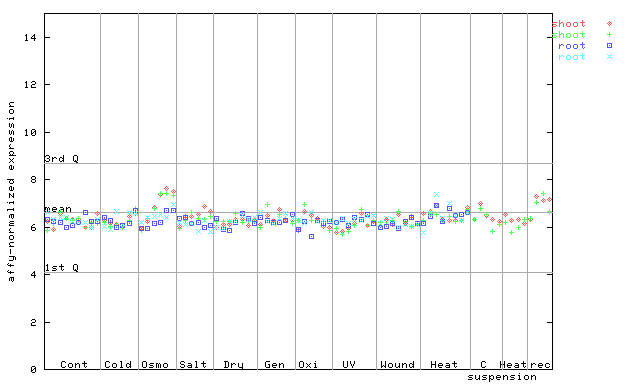
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
260677_at
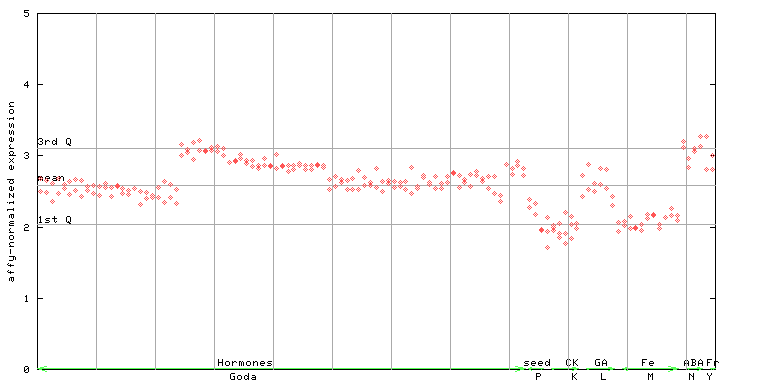
X axis is samples (xls file), and Y axis is log-expression.
|












