| functional annotation |
| Function |
chloroplast RNA binding protein |


|
| GO BP |
|
GO:0033499 [list] [network] galactose catabolic process via UDP-galactose
|
(14 genes)
|
IBA
|
 |
|
GO:0019388 [list] [network] galactose catabolic process
|
(16 genes)
|
IBA
|
 |
|
GO:0032544 [list] [network] plastid translation
|
(16 genes)
|
IMP
|
 |
|
GO:0045727 [list] [network] positive regulation of translation
|
(26 genes)
|
IMP
|
 |
|
GO:0042631 [list] [network] cellular response to water deprivation
|
(35 genes)
|
IEP
|
 |
|
GO:0007623 [list] [network] circadian rhythm
|
(111 genes)
|
IEP
IMP
|
 |
|
GO:0005996 [list] [network] monosaccharide metabolic process
|
(150 genes)
|
IMP
|
 |
|
GO:0000272 [list] [network] polysaccharide catabolic process
|
(170 genes)
|
IEA
|
 |
|
GO:0009658 [list] [network] chloroplast organization
|
(194 genes)
|
IMP
|
 |
|
GO:0009611 [list] [network] response to wounding
|
(212 genes)
|
IEP
|
 |
|
GO:0006364 [list] [network] rRNA processing
|
(276 genes)
|
IGI
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(399 genes)
|
IEP
|
 |
|
GO:0009409 [list] [network] response to cold
|
(411 genes)
|
IEP
|
 |
|
GO:0045893 [list] [network] positive regulation of transcription, DNA-templated
|
(495 genes)
|
IMP
|
 |
|
GO:0010468 [list] [network] regulation of gene expression
|
(2484 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001322308.1
NP_172405.1
|
| BLAST |
NP_001322308.1
NP_172405.1
|
| Orthologous |
[Ortholog page]
LOC4352083 (osa)
LOC7478886 (ppo)
LOC25499634 (mtr)
LOC100193292 (zma)
LOC100261308 (vvi)
LOC100791076 (gma)
LOC100799721 (gma)
LOC101253094 (sly)
LOC103871660 (bra)
|
Subcellular
localization
wolf |
|
nucl 3,
cyto 3,
cyto_nucl 3,
chlo 2,
cyto_pero 2
|
(predict for NP_001322308.1)
|
|
chlo 7,
nucl 1,
vacu 1,
pero 1,
cyto_nucl 1,
cysk_nucl 1,
nucl_plas 1
|
(predict for NP_172405.1)
|
|
Subcellular
localization
TargetP |
|
other 7,
scret 3
|
(predict for NP_001322308.1)
|
|
other 3
|
(predict for NP_172405.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath01200 |
Carbon metabolism |
8 |
 |
| ath00630 |
Glyoxylate and dicarboxylate metabolism |
7 |
 |
| ath00910 |
Nitrogen metabolism |
4 |
 |
| ath00710 |
Carbon fixation in photosynthetic organisms |
4 |
 |
| ath00260 |
Glycine, serine and threonine metabolism |
3 |
 |
Genes directly connected with CRB on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 16.0 |
SBPASE |
sedoheptulose-bisphosphatase |
[detail] |
824746 |
| 13.1 |
SHM1 |
serine transhydroxymethyltransferase 1 |
[detail] |
829949 |
| 12.9 |
GAPB |
glyceraldehyde-3-phosphate dehydrogenase B subunit |
[detail] |
840895 |
| 12.0 |
ZKT |
protein containing PDZ domain, a K-box domain, and a TPR region |
[detail] |
841995 |
| 11.7 |
PMDH2 |
peroxisomal NAD-malate dehydrogenase 2 |
[detail] |
830825 |
| 11.2 |
CA1 |
carbonic anhydrase 1 |
[detail] |
821134 |
| 11.1 |
LrgB |
membrane protein |
[detail] |
840100 |
| 10.5 |
AT3G48420 |
Haloacid dehalogenase-like hydrolase (HAD) superfamily protein |
[detail] |
824000 |
| 9.8 |
GLU1 |
glutamate synthase 1 |
[detail] |
830292 |
| 9.1 |
GS2 |
glutamine synthetase 2 |
[detail] |
833535 |
| 8.7 |
HCF101 |
ATP binding protein |
[detail] |
822033 |
| 8.2 |
KEA1 |
K+ efflux antiporter 1 |
[detail] |
837332 |
| 8.0 |
AT1G16080 |
nuclear protein |
[detail] |
838178 |
| 7.9 |
GLDP1 |
glycine decarboxylase P-protein 1 |
[detail] |
829438 |
| 7.2 |
AT1G71500 |
Rieske (2Fe-2S) domain-containing protein |
[detail] |
843491 |
| 5.6 |
LHCB4.3 |
light harvesting complex photosystem II |
[detail] |
818599 |
|
Coexpressed
gene list |
[Coexpressed gene list for CRB]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
263676_at
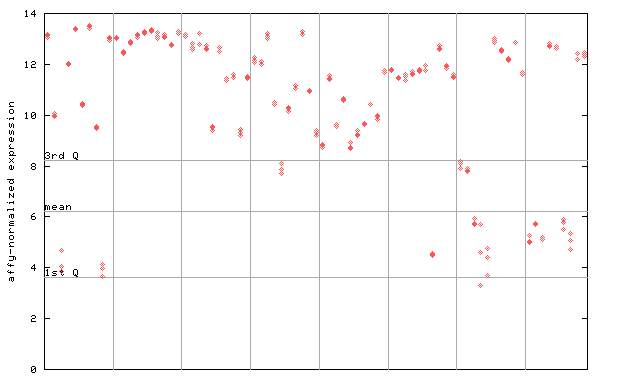
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
263676_at
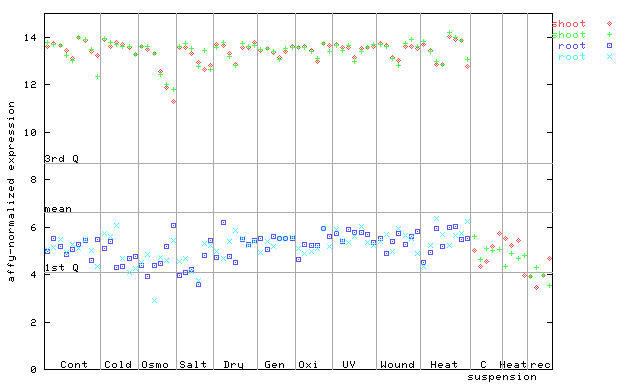
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
263676_at
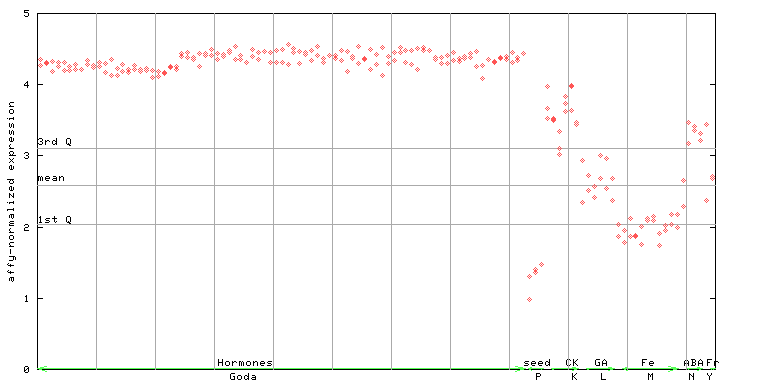
X axis is samples (xls file), and Y axis is log-expression.
|















