| functional annotation |
| Function |
UDP-Glycosyltransferase superfamily protein |




|
| GO BP |
|
GO:0019252 [list] [network] starch biosynthetic process
|
(25 genes)
|
IEA
|
 |
|
GO:0071478 [list] [network] cellular response to radiation
|
(166 genes)
|
IEA
|
 |
|
GO:0042440 [list] [network] pigment metabolic process
|
(230 genes)
|
IEA
|
 |
|
GO:0009639 [list] [network] response to red or far red light
|
(365 genes)
|
IEA
|
 |
|
GO:0009409 [list] [network] response to cold
|
(472 genes)
|
IEA
|
 |
|
GO:0046394 [list] [network] carboxylic acid biosynthetic process
|
(517 genes)
|
IEA
|
 |
|
GO:1901135 [list] [network] carbohydrate derivative metabolic process
|
(589 genes)
|
IEA
|
 |
|
GO:0009753 [list] [network] response to jasmonic acid
|
(611 genes)
|
IEA
|
 |
|
GO:0008610 [list] [network] lipid biosynthetic process
|
(657 genes)
|
IEA
|
 |
|
GO:0019748 [list] [network] secondary metabolic process
|
(766 genes)
|
IEA
|
 |
|
GO:0044255 [list] [network] cellular lipid metabolic process
|
(822 genes)
|
IEA
|
 |
|
GO:0010035 [list] [network] response to inorganic substance
|
(2100 genes)
|
IEA
|
 |
= more 7 terms =
|
| GO CC |
|
| GO MF |
|
GO:0102502 [list] [network] ADP-glucose-starch glucosyltransferase activity
|
(1 genes)
|
IEA
|
 |
|
GO:0004373 [list] [network] glycogen (starch) synthase activity
|
(5 genes)
|
IEA
|
 |
|
GO:0016757 [list] [network] glycosyltransferase activity
|
(567 genes)
|
ISS
|
 |
|
GO:0005515 [list] [network] protein binding
|
(5066 genes)
|
IPI
|
 |
|
| KEGG |
ath00500 [list] [network] Starch and sucrose metabolism (172 genes) |
 |
| Protein |
NP_174566.1
|
| BLAST |
NP_174566.1
|
| Orthologous |
[Ortholog page]
LOC542837 (tae)
LOC543484 (tae)
LOC4340018 (osa)
LOC4343010 (osa)
LOC7455034 (ppo)
LOC7491408 (ppo)
LOC25484575 (mtr)
LOC25500617 (mtr)
LOC100037476 (gma)
LOC100037526 (tae)
GBSSIa (gma)
LOC100787590 (gma)
LOC100817424 (gma)
GBSS1 (sly)
LOC103874772 (bra)
LOC123053932 (tae)
LOC123189823 (tae)
LOC123412237 (hvu)
LOC123427313 (hvu)
|
Subcellular
localization
wolf |
|
chlo 9
|
(predict for NP_174566.1)
|
|
Subcellular
localization
TargetP |
|
chlo 9
|
(predict for NP_174566.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00500 |
Starch and sucrose metabolism |
3 |
 |
| ath00520 |
Amino sugar and nucleotide sugar metabolism |
2 |
 |
| ath01250 |
Biosynthesis of nucleotide sugars |
2 |
 |
| ath00562 |
Inositol phosphate metabolism |
2 |
 |
Genes directly connected with GBSS1 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 7.8 |
AT1G80130 |
Tetratricopeptide repeat (TPR)-like superfamily protein |
[detail] |
844353 |
| 6.4 |
NDA1 |
alternative NAD(P)H dehydrogenase 1 |
[detail] |
837229 |
| 6.4 |
G-TMT |
gamma-tocopherol methyltransferase |
[detail] |
842805 |
| 5.6 |
ARC5 |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
821509 |
| 5.5 |
MIPS2 |
myo-inositol-1-phosphate synthase 2 |
[detail] |
816757 |
| 5.3 |
APL4 |
Glucose-1-phosphate adenylyltransferase family protein |
[detail] |
816697 |
| 4.8 |
AT5G20190 |
Tetratricopeptide repeat (TPR)-like superfamily protein |
[detail] |
832142 |
| 4.6 |
CXE13 |
carboxyesterase 13 |
[detail] |
824031 |
|
Coexpressed
gene list |
[Coexpressed gene list for GBSS1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
261191_at
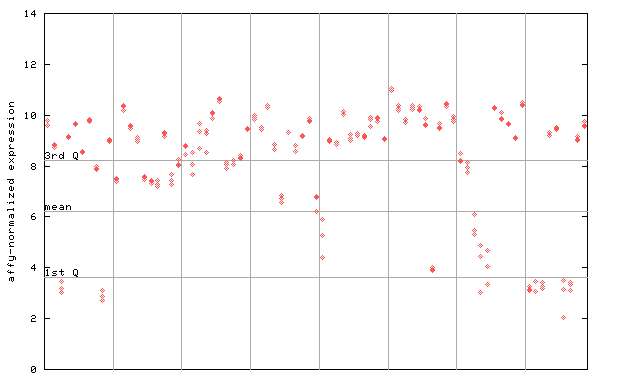
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
261191_at
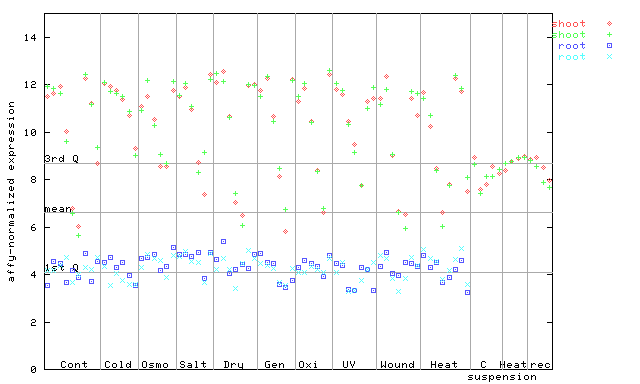
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
261191_at
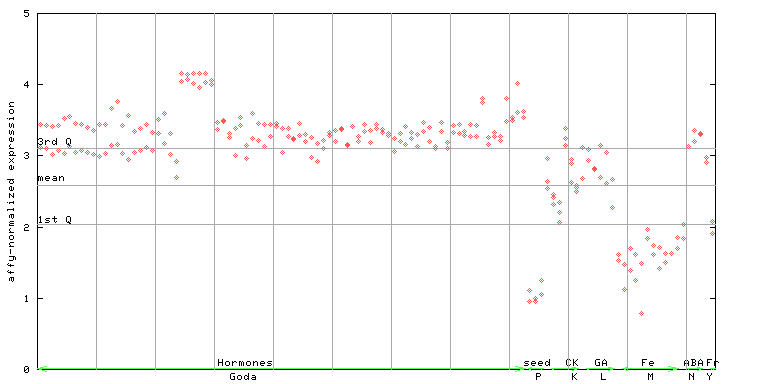
X axis is samples (xls file), and Y axis is log-expression.
|
















