| functional annotation |
| Function |
leucine-rich repeat transmembrane protein kinase family protein |




|
| GO BP |
|
GO:1900426 [list] [network] positive regulation of defense response to bacterium
|
(31 genes)
|
IEA
|
 |
|
GO:0002253 [list] [network] activation of immune response
|
(55 genes)
|
IEA
|
 |
|
GO:0002239 [list] [network] response to oomycetes
|
(72 genes)
|
IEA
|
 |
|
GO:0000302 [list] [network] response to reactive oxygen species
|
(141 genes)
|
IEA
|
 |
|
GO:0045087 [list] [network] innate immune response
|
(161 genes)
|
IEA
|
 |
|
GO:0010150 [list] [network] leaf senescence
|
(195 genes)
|
IEA
|
 |
|
GO:0071407 [list] [network] cellular response to organic cyclic compound
|
(250 genes)
|
IEA
|
 |
|
GO:0048366 [list] [network] leaf development
|
(465 genes)
|
IEA
|
 |
|
GO:0006979 [list] [network] response to oxidative stress
|
(545 genes)
|
IEA
|
 |
|
GO:0010243 [list] [network] response to organonitrogen compound
|
(591 genes)
|
IEA
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(607 genes)
|
IEA
|
 |
|
GO:0006468 [list] [network] protein phosphorylation
|
(639 genes)
|
IEA
|
 |
|
GO:0050832 [list] [network] defense response to fungus
|
(705 genes)
|
IEA
|
 |
|
GO:0031347 [list] [network] regulation of defense response
|
(749 genes)
|
IEA
|
 |
|
GO:0009611 [list] [network] response to wounding
|
(816 genes)
|
IEA
|
 |
|
GO:0009755 [list] [network] hormone-mediated signaling pathway
|
(904 genes)
|
IEA
|
 |
|
GO:0009414 [list] [network] response to water deprivation
|
(1006 genes)
|
IEA
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(1086 genes)
|
IEA
|
 |
|
GO:1901701 [list] [network] cellular response to oxygen-containing compound
|
(1115 genes)
|
IEA
|
 |
|
GO:0098542 [list] [network] defense response to other organism
|
(2060 genes)
|
IEA
|
 |
|
| GO CC |
|
GO:0005886 [list] [network] plasma membrane
|
(2529 genes)
|
ISM
|
 |
|
| GO MF |
|
GO:0005524 [list] [network] ATP binding
|
(253 genes)
|
IEA
|
 |
|
GO:0004672 [list] [network] protein kinase activity
|
(768 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_174702.1
|
| BLAST |
NP_174702.1
|
| Orthologous |
[Ortholog page]
LOC4351679 (osa)
LOC7467429 (ppo)
LOC7467430 (ppo)
LOC18104471 (ppo)
LOC25484960 (mtr)
LOC100796118 (gma)
LOC100796647 (gma)
LOC101264085 (sly)
LOC101264781 (sly)
LOC102669972 (gma)
LOC103833596 (bra)
LOC123091327 (tae)
LOC123111016 (tae)
LOC123120045 (tae)
LOC123125687 (tae)
LOC123395724 (hvu)
LOC123413218 (hvu)
LOC123447917 (hvu)
|
Subcellular
localization
wolf |
|
plas 6,
vacu 1,
nucl 1,
mito 1,
extr 1,
E.R. 1
|
(predict for NP_174702.1)
|
|
Subcellular
localization
TargetP |
|
other 5,
scret 3
|
(predict for NP_174702.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04626 |
Plant-pathogen interaction |
2 |
 |
Genes directly connected with AT1G34420 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 5.9 |
EDS1 |
alpha/beta-Hydrolases superfamily protein |
[detail] |
823964 |
| 5.8 |
AT1G66880 |
Protein kinase superfamily protein |
[detail] |
843006 |
| 5.8 |
AT3G48080 |
alpha/beta-Hydrolases superfamily protein |
[detail] |
823963 |
|
Coexpressed
gene list |
[Coexpressed gene list for AT1G34420]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
261161_at
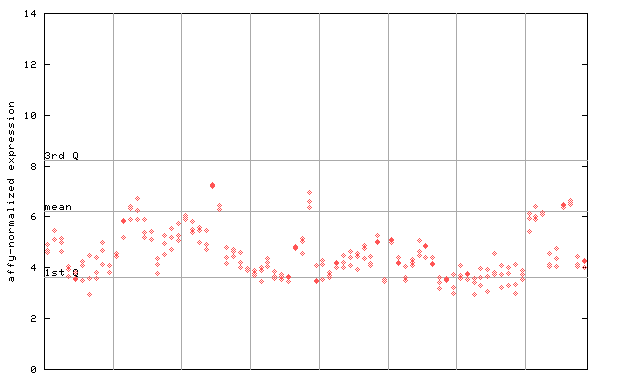
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
261161_at
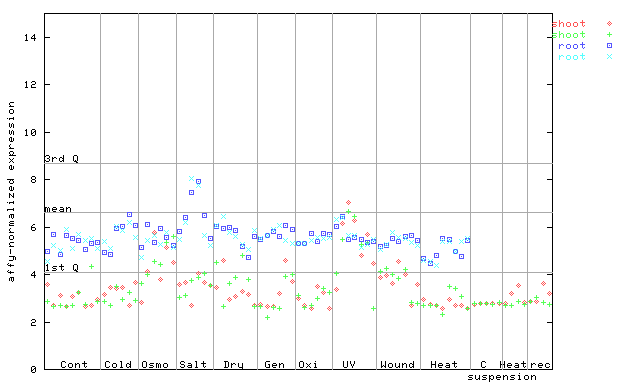
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
261161_at
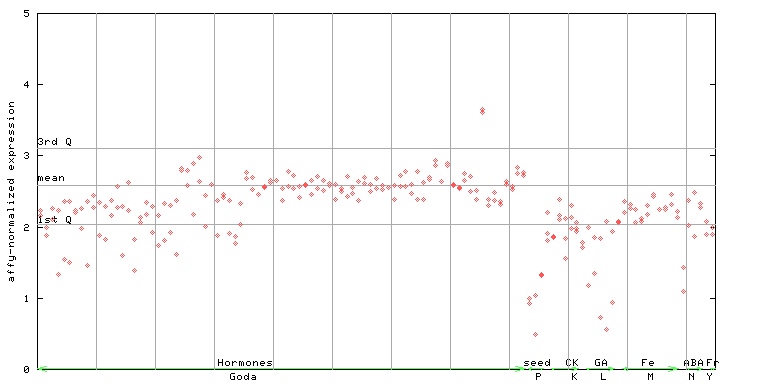
X axis is samples (xls file), and Y axis is log-expression.
|












