| functional annotation |
| Function |
Signal transduction histidine kinase, hybrid-type, ethylene sensor |




|
| GO BP |
|
GO:0009727 [list] [network] detection of ethylene stimulus
|
(3 genes)
|
IMP
|
 |
|
GO:0009871 [list] [network] jasmonic acid and ethylene-dependent systemic resistance, ethylene mediated signaling pathway
|
(4 genes)
|
TAS
|
 |
|
GO:0050665 [list] [network] hydrogen peroxide biosynthetic process
|
(9 genes)
|
IMP
|
 |
|
GO:0052544 [list] [network] defense response by callose deposition in cell wall
|
(16 genes)
|
IMP
|
 |
|
GO:0010105 [list] [network] negative regulation of ethylene-activated signaling pathway
|
(20 genes)
|
TAS
|
 |
|
GO:0009625 [list] [network] response to insect
|
(33 genes)
|
IMP
|
 |
|
GO:0009690 [list] [network] cytokinin metabolic process
|
(38 genes)
|
IMP
|
 |
|
GO:0010182 [list] [network] sugar mediated signaling pathway
|
(38 genes)
|
TAS
|
 |
|
GO:0010162 [list] [network] seed dormancy process
|
(49 genes)
|
IMP
|
 |
|
GO:1900140 [list] [network] regulation of seedling development
|
(98 genes)
|
IMP
|
 |
|
GO:0010119 [list] [network] regulation of stomatal movement
|
(105 genes)
|
IMP
|
 |
|
GO:0009739 [list] [network] response to gibberellin
|
(116 genes)
|
IMP
|
 |
|
GO:0002237 [list] [network] response to molecule of bacterial origin
|
(118 genes)
|
IMP
|
 |
|
GO:0010087 [list] [network] phloem or xylem histogenesis
|
(158 genes)
|
IMP
|
 |
|
GO:0009723 [list] [network] response to ethylene
|
(241 genes)
|
IMP
|
 |
|
GO:0009408 [list] [network] response to heat
|
(287 genes)
|
IMP
|
 |
|
GO:0009733 [list] [network] response to auxin
|
(384 genes)
|
IMP
|
 |
|
GO:0051301 [list] [network] cell division
|
(442 genes)
|
IMP
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(607 genes)
|
IEP
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(980 genes)
|
IMP
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(1086 genes)
|
IMP
|
 |
|
GO:0006952 [list] [network] defense response
|
(2370 genes)
|
TAS
|
 |
|
| GO CC |
|
GO:0005789 [list] [network] endoplasmic reticulum membrane
|
(108 genes)
|
IDA
|
 |
|
GO:0005783 [list] [network] endoplasmic reticulum
|
(856 genes)
|
IDA
|
 |
|
| GO MF |
|
GO:0038199 [list] [network] ethylene receptor activity
|
(3 genes)
|
IMP
|
 |
|
GO:0051740 [list] [network] ethylene binding
|
(5 genes)
|
IDA
|
 |
|
GO:0000155 [list] [network] phosphorelay sensor kinase activity
|
(14 genes)
|
IEA
|
 |
|
GO:0004673 [list] [network] protein histidine kinase activity
|
(17 genes)
|
IMP
ISS
TAS
|
 |
|
GO:0042802 [list] [network] identical protein binding
|
(404 genes)
|
IPI
|
 |
|
GO:0005515 [list] [network] protein binding
|
(5066 genes)
|
IPI
|
 |
|
| KEGG |
ath04016 [list] [network] MAPK signaling pathway - plant (139 genes) |
 |
| ath04075 [list] [network] Plant hormone signal transduction (291 genes) |
 |
| Protein |
NP_176808.3
|
| BLAST |
NP_176808.3
|
| Orthologous |
[Ortholog page]
ETR1 (sly)
ETR2 (sly)
LOC7470545 (ppo)
LOC11434695 (mtr)
LOC18096706 (ppo)
LOC100795340 (gma)
LOC100795799 (gma)
ETR1 (bra)
|
Subcellular
localization
wolf |
|
cyto 6,
plas 2,
nucl 1,
E.R. 1,
cysk 1,
cysk_nucl 1
|
(predict for NP_176808.3)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_176808.3)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04075 |
Plant hormone signal transduction |
3 |
 |
| ath03015 |
mRNA surveillance pathway |
2 |
 |
| ath04016 |
MAPK signaling pathway - plant |
2 |
 |
Genes directly connected with ETR1 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 6.2 |
UBP16 |
ubiquitin-specific protease 16 |
[detail] |
828558 |
| 6.2 |
BAK1 |
BRI1-associated receptor kinase |
[detail] |
829480 |
| 6.0 |
LRS1 |
Transducin/WD40 repeat-like superfamily protein |
[detail] |
819671 |
| 5.3 |
AT5G11850 |
Protein kinase superfamily protein |
[detail] |
831058 |
|
Coexpressed
gene list |
[Coexpressed gene list for ETR1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
260133_at
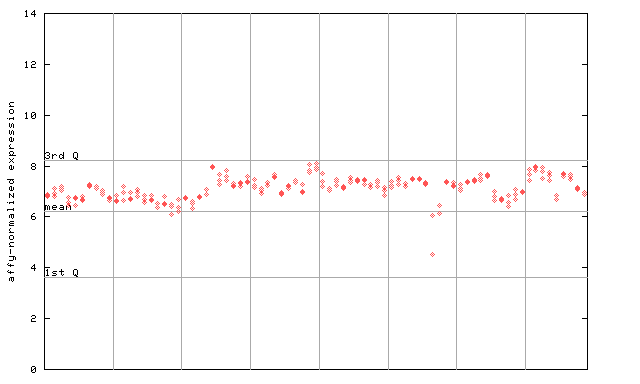
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
260133_at
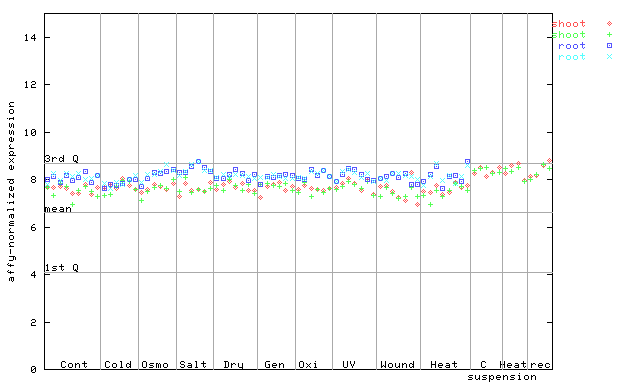
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
260133_at
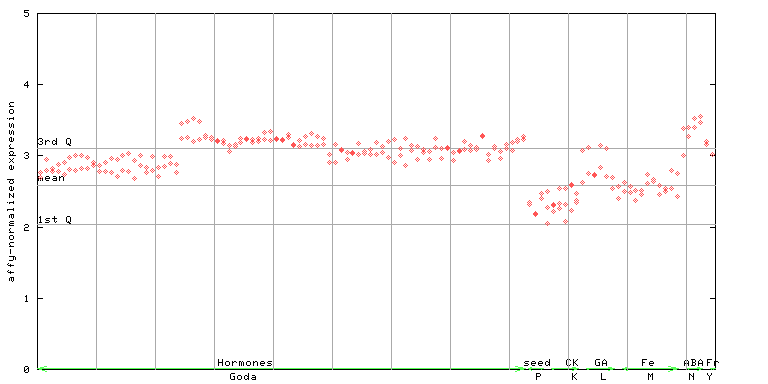
X axis is samples (xls file), and Y axis is log-expression.
|
















