[←][→] ath
| functional annotation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | Protein kinase superfamily protein |




|
||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_176871.2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_176871.2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] AT5G38260 (ath) AT1G66910 (ath) AT1G66920 (ath) AT1G66930 (ath) AT1G66940 (ath) AT1G67025 (ath) LOC4325673 (osa) LOC4325680 (osa) LOC4327950 (osa) LOC7475645 (ppo) LOC7477440 (ppo) LOC7493513 (ppo) LOC11426496 (mtr) LOC11429097 (mtr) LOC11432157 (mtr) LOC11439933 (mtr) LOC11440978 (mtr) LOC11442395 (mtr) LOC11443712 (mtr) LOC11444473 (mtr) LOC11445504 (mtr) LOC18097554 (ppo) LOC18099461 (ppo) LOC18099463 (ppo) LOC18099466 (ppo) LOC18099467 (ppo) LOC18099468 (ppo) LOC18099470 (ppo) LOC18099472 (ppo) LOC18109660 (ppo) LOC18109684 (ppo) LOC18109711 (ppo) LOC18109713 (ppo) LOC18110242 (ppo) LOC18110245 (ppo) LOC25485727 (mtr) LOC25496801 (mtr) LOC25496804 (mtr) LOC25496805 (mtr) LOC25496811 (mtr) LOC25496829 (mtr) LOC25496836 (mtr) LOC100778621 (gma) LOC100781427 (gma) LOC100781977 (gma) LOC100783861 (gma) LOC100784387 (gma) LOC100785450 (gma) LOC100785979 (gma) LOC100786495 (gma) LOC100787576 (gma) LOC100792936 (gma) LOC100793451 (gma) LOC100794505 (gma) LOC100796618 (gma) LOC100797690 (gma) LOC100798745 (gma) LOC100799268 (gma) LOC100799800 (gma) LOC100800335 (gma) LOC100807312 (gma) LOC100809508 (gma) LOC100816285 (gma) LOC101243709 (sly) LOC101261220 (sly) LOC101263961 (sly) LOC102665158 (gma) LOC103831142 (bra) LOC103834317 (bra) LOC103840859 (bra) LOC103850109 (bra) LOC103852346 (bra) LOC103852351 (bra) LOC103852354 (bra) LOC103852357 (bra) LOC103852359 (bra) LOC103864029 (bra) LOC106794113 (gma) LOC106799440 (gma) LOC107276391 (osa) LOC107278584 (osa) LOC107281842 (osa) LOC112324599 (ppo) LOC112325668 (ppo) LOC112325671 (ppo) LOC112937635 (osa) LOC112937810 (osa) LOC112940885 (sly) LOC117125698 (bra) LOC117132650 (bra) LOC120577941 (mtr) LOC121172853 (gma) LOC121174286 (gma) LOC123056669 (tae) LOC123057696 (tae) LOC123057697 (tae) LOC123059906 (tae) LOC123059991 (tae) LOC123059992 (tae) LOC123061825 (tae) LOC123061831 (tae) LOC123064603 (tae) LOC123064913 (tae) LOC123067230 (tae) LOC123067233 (tae) LOC123067237 (tae) LOC123067242 (tae) LOC123067243 (tae) LOC123068429 (tae) LOC123068430 (tae) LOC123068431 (tae) LOC123068432 (tae) LOC123068433 (tae) LOC123068435 (tae) LOC123068436 (tae) LOC123068438 (tae) LOC123068443 (tae) LOC123068444 (tae) LOC123068446 (tae) LOC123068447 (tae) LOC123068448 (tae) LOC123068450 (tae) LOC123068451 (tae) LOC123068453 (tae) LOC123068454 (tae) LOC123068455 (tae) LOC123070520 (tae) LOC123073838 (tae) LOC123076063 (tae) LOC123076066 (tae) LOC123076940 (tae) LOC123076950 (tae) LOC123076958 (tae) LOC123076959 (tae) LOC123076962 (tae) LOC123076963 (tae) LOC123078931 (tae) LOC123101195 (tae) LOC123106542 (tae) LOC123131548 (tae) LOC123133048 (tae) LOC123142898 (tae) LOC123161850 (tae) LOC123169531 (tae) LOC123170917 (tae) LOC123441359 (hvu) LOC123441360 (hvu) LOC123441362 (hvu) LOC123441372 (hvu) LOC123442547 (hvu) LOC123442576 (hvu) LOC123442577 (hvu) LOC123442578 (hvu) LOC123442581 (hvu) LOC123442582 (hvu) LOC123444154 (hvu) | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for AT1G67000] | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | |||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
255879_at
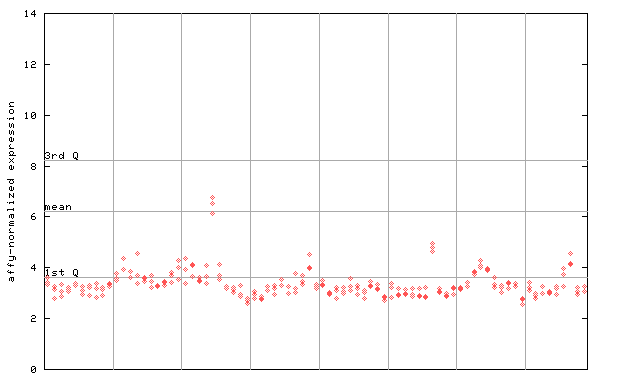
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
255879_at
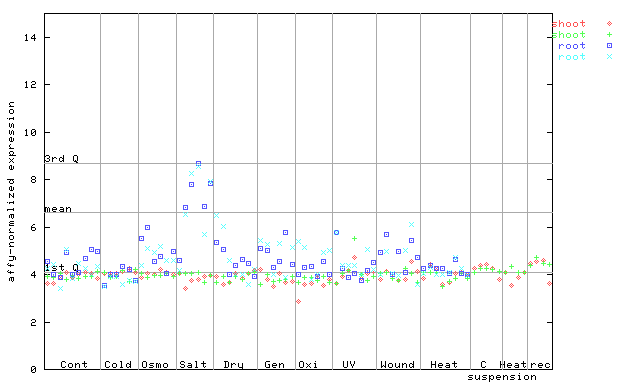
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
255879_at
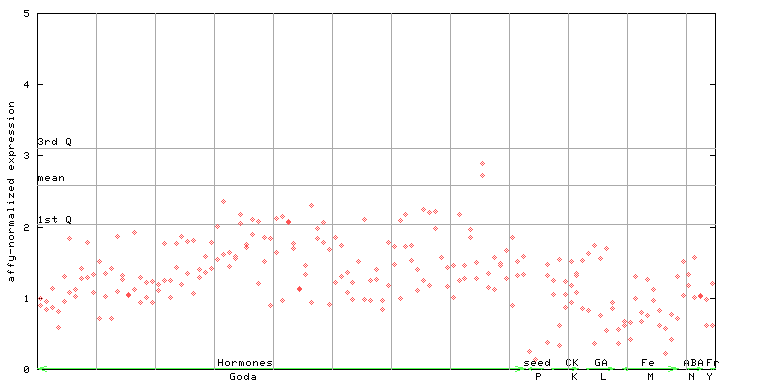
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 843018 |


|
| Refseq ID (protein) | NP_176871.2 |  |
The preparation time of this page was 0.1 [sec].




