| functional annotation |
| Function |
K-box region and MADS-box transcription factor family protein |


|
| GO BP |
|
GO:0010076 [list] [network] maintenance of floral meristem identity
|
(9 genes)
|
IGI
|
 |
|
GO:0010582 [list] [network] floral meristem determinacy
|
(20 genes)
|
IBA
IGI
|
 |
|
GO:0009933 [list] [network] meristem structural organization
|
(54 genes)
|
IMP
|
 |
|
GO:0045944 [list] [network] positive regulation of transcription by RNA polymerase II
|
(206 genes)
|
IEA
|
 |
|
GO:0009908 [list] [network] flower development
|
(447 genes)
|
IMP
|
 |
|
GO:0045893 [list] [network] positive regulation of transcription, DNA-templated
|
(495 genes)
|
IDA
IGI
|
 |
|
GO:0030154 [list] [network] cell differentiation
|
(695 genes)
|
IEA
|
 |
|
GO:0007275 [list] [network] multicellular organism development
|
(2714 genes)
|
IBA
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0046982 [list] [network] protein heterodimerization activity
|
(122 genes)
|
IPI
|
 |
|
GO:0000977 [list] [network] RNA polymerase II regulatory region sequence-specific DNA binding
|
(152 genes)
|
IBA
|
 |
|
GO:0008134 [list] [network] transcription factor binding
|
(173 genes)
|
IBA
|
 |
|
GO:0043565 [list] [network] sequence-specific DNA binding
|
(796 genes)
|
IBA
|
 |
|
GO:0044212 [list] [network] transcription regulatory region DNA binding
|
(871 genes)
|
IBA
|
 |
|
GO:0003700 [list] [network] DNA-binding transcription factor activity
|
(1599 genes)
|
ISS
|
 |
|
GO:0003677 [list] [network] DNA binding
|
(2280 genes)
|
IDA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001320361.1
NP_001320362.1
NP_177074.1
|
| BLAST |
NP_001320361.1
NP_001320362.1
NP_177074.1
|
| Orthologous |
[Ortholog page]
LOC542032 (zma)
LOC542041 (zma)
LOC542045 (zma)
MADS-MC (sly)
FUL1 (sly)
FUL2 (sly)
AGL8 (ath)
CAL (ath)
LOC4334140 (osa)
LOC4342214 (osa)
LOC4343851 (osa)
LOC7468138 (ppo)
LOC7471072 (ppo)
LOC11418521 (mtr)
LOC11435341 (mtr)
LOC25488419 (mtr)
LOC25493907 (mtr)
AP1 (vvi)
VFUL-L (vvi)
LOC100257072 (vvi)
LOC100783045 (gma)
LOC100784742 (gma)
FULC (gma)
LOC100805348 (gma)
AP1 (gma)
LOC100811588 (gma)
AP1A (gma)
LOC100819745 (gma)
LOC103830938 (bra)
LOC103831316 (bra)
LOC103835484 (bra)
LOC103852552 (bra)
LOC103854924 (bra)
LOC103860551 (bra)
MBP20 (sly)
|
Subcellular
localization
wolf |
|
cyto 4,
nucl 3,
cyto_E.R. 2,
pero 1,
nucl_plas 1
|
(predict for NP_001320361.1)
|
|
nucl 10
|
(predict for NP_001320362.1)
|
|
nucl 10
|
(predict for NP_177074.1)
|
|
Subcellular
localization
TargetP |
|
other 7
|
(predict for NP_001320361.1)
|
|
mito 7,
other 5
|
(predict for NP_001320362.1)
|
|
mito 7,
other 5
|
(predict for NP_177074.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with AP1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 12.9 |
RXF26 |
GDSL-like Lipase/Acylhydrolase family protein |
[detail] |
842212 |
| 12.7 |
AT5G22430 |
Pollen Ole e 1 allergen and extensin family protein |
[detail] |
832304 |
| 12.3 |
AT5G44630 |
Terpenoid cyclases/Protein prenyltransferases superfamily protein |
[detail] |
834491 |
| 10.6 |
SEP2 |
K-box region and MADS-box transcription factor family protein |
[detail] |
821151 |
| 10.1 |
CYP706A3 |
cytochrome P450, family 706, subfamily A, polypeptide 3 |
[detail] |
834490 |
| 9.0 |
SHN1 |
Integrase-type DNA-binding superfamily protein |
[detail] |
838105 |
| 8.7 |
SEP1 |
K-box region and MADS-box transcription factor family protein |
[detail] |
831436 |
| 8.2 |
AP3 |
K-box region and MADS-box transcription factor family protein |
[detail] |
824601 |
| 7.8 |
AGL8 |
AGAMOUS-like 8 |
[detail] |
836212 |
| 7.5 |
PI |
K-box region and MADS-box transcription factor family protein |
[detail] |
832146 |
| 7.1 |
REM1 |
Transcriptional factor B3 family protein |
[detail] |
829288 |
| 5.9 |
AT4G34400 |
AP2/B3-like transcriptional factor family protein |
[detail] |
829590 |
| 5.1 |
AT3G28500 |
60S acidic ribosomal protein family |
[detail] |
822480 |
| 5.1 |
FPF1 |
flowering promoting factor 1 |
[detail] |
832555 |
| 4.8 |
CYP96A9 |
cytochrome P450, family 96, subfamily A, polypeptide 9 |
[detail] |
830103 |
| 3.8 |
FAF2 |
FANTASTIC four-like protein (DUF3049) |
[detail] |
839563 |
|
Coexpressed
gene list |
[Coexpressed gene list for AP1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
259372_at
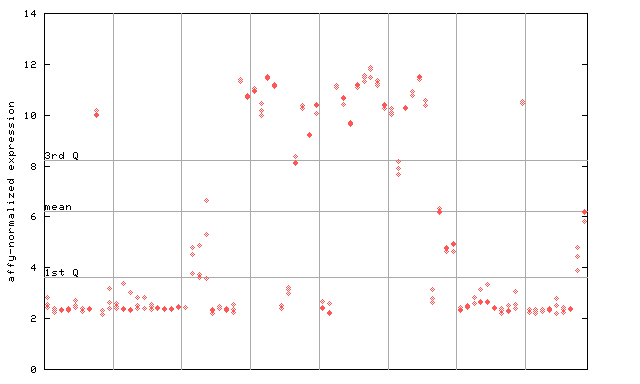
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
259372_at
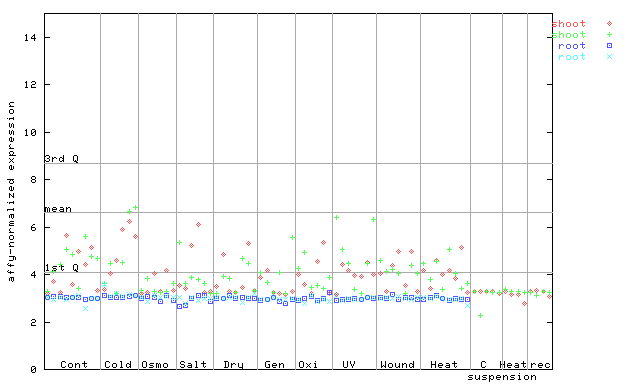
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
259372_at
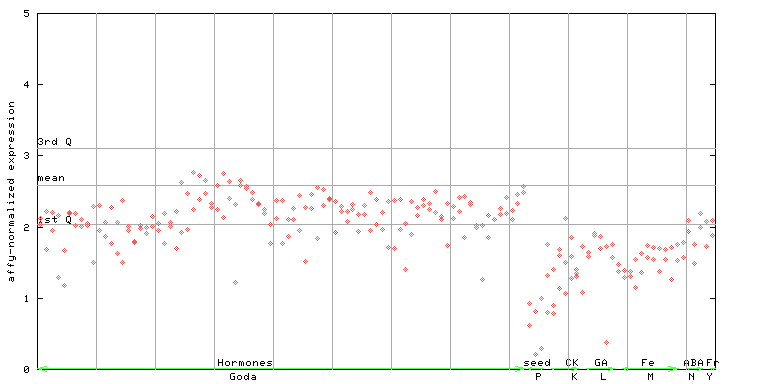
X axis is samples (xls file), and Y axis is log-expression.
|











