[←][→] ath
| functional annotation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | Phosphoglucomutase/phosphomannomutase family protein |


|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | ath00010 [list] [network] Glycolysis / Gluconeogenesis (119 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00030 [list] [network] Pentose phosphate pathway (58 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00052 [list] [network] Galactose metabolism (57 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00230 [list] [network] Purine metabolism (87 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00500 [list] [network] Starch and sucrose metabolism (172 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00520 [list] [network] Amino sugar and nucleotide sugar metabolism (133 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath01250 [list] [network] Biosynthesis of nucleotide sugars (100 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_001154464.1 NP_001154465.1 NP_177230.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_001154464.1 NP_001154465.1 NP_177230.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] LOC543348 (tae) PGM3 (ath) LOC4333895 (osa) LOC7475519 (ppo) LOC7482898 (ppo) LOC11413693 (mtr) LOC100780166 (gma) LOC100802789 (gma) LOC101267885 (sly) LOC103835677 (bra) LOC103840790 (bra) LOC123087100 (tae) LOC123447463 (hvu) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for PGM2] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
260207_at
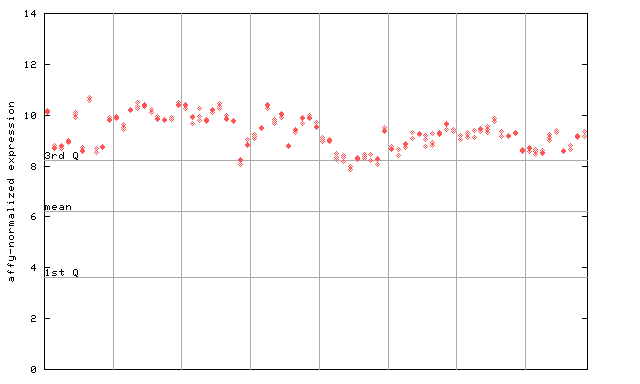
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
260207_at
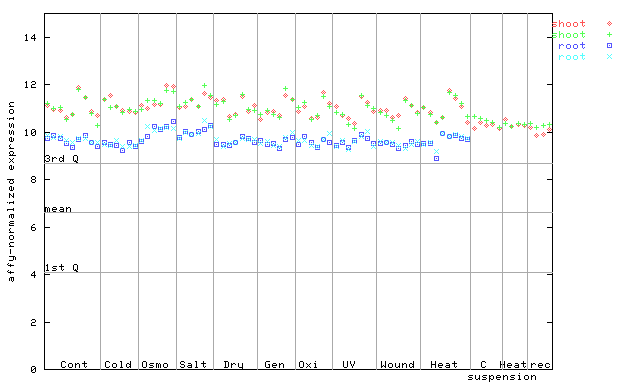
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
260207_at
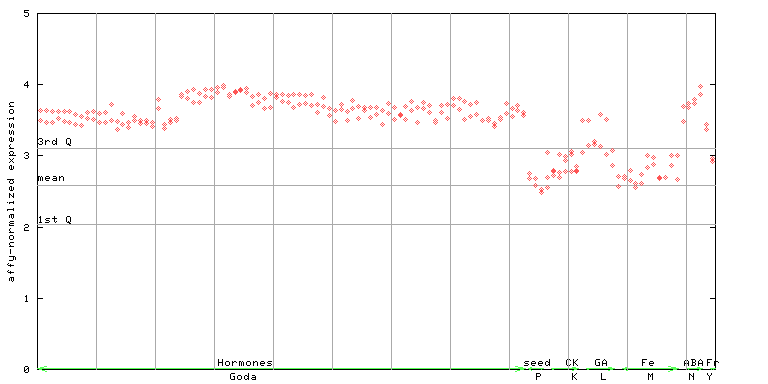
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 843410 |


|
| Refseq ID (protein) | NP_001154464.1 |  |
| NP_001154465.1 |  |
|
| NP_177230.1 |  |
|
The preparation time of this page was 0.2 [sec].






