| functional annotation |
| Function |
Protein kinase superfamily protein |


|
| GO BP |
|
GO:1902171 [list] [network] regulation of tocopherol cyclase activity
|
(2 genes)
|
IMP
|
 |
|
GO:0080177 [list] [network] plastoglobule organization
|
(3 genes)
|
IMP
|
 |
|
GO:0080183 [list] [network] response to photooxidative stress
|
(11 genes)
|
IMP
|
 |
|
GO:0050821 [list] [network] protein stabilization
|
(20 genes)
|
IMP
|
 |
|
GO:0006995 [list] [network] cellular response to nitrogen starvation
|
(33 genes)
|
IMP
|
 |
|
GO:0009644 [list] [network] response to high light intensity
|
(65 genes)
|
IMP
|
 |
|
GO:0010114 [list] [network] response to red light
|
(74 genes)
|
IMP
|
 |
|
GO:0009658 [list] [network] chloroplast organization
|
(194 genes)
|
IMP
|
 |
|
GO:0009414 [list] [network] response to water deprivation
|
(361 genes)
|
IMP
|
 |
|
GO:0006468 [list] [network] protein phosphorylation
|
(921 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_565214.1
|
| BLAST |
NP_565214.1
|
| Orthologous |
[Ortholog page]
LOC4338416 (osa)
LOC7474529 (ppo)
LOC11423084 (mtr)
LOC25480022 (mtr)
LOC100253506 (vvi)
LOC100286057 (zma)
LOC100802848 (gma)
LOC100803551 (gma)
LOC100804500 (gma)
ABC1K3 (sly)
LOC103630188 (zma)
LOC103830474 (bra)
LOC103853454 (bra)
|
Subcellular
localization
wolf |
|
chlo 7,
cyto 1,
extr 1,
vacu 1,
E.R. 1,
E.R._vacu 1,
cyto_E.R. 1
|
(predict for NP_565214.1)
|
|
Subcellular
localization
TargetP |
|
chlo 7
|
(predict for NP_565214.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00906 |
Carotenoid biosynthesis |
2 |
 |
Genes directly connected with AT1G79600 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 8.4 |
PDS3 |
phytoene desaturase 3 |
[detail] |
827061 |
| 8.2 |
VAR2 |
FtsH extracellular protease family |
[detail] |
817646 |
| 7.9 |
ABA1 |
zeaxanthin epoxidase (ZEP) (ABA1) |
[detail] |
836838 |
| 7.7 |
AT3G07700 |
Protein kinase superfamily protein |
[detail] |
819961 |
| 7.0 |
AT1G50020 |
tubulin alpha-6 chain |
[detail] |
841426 |
| 6.7 |
TIC55-II |
translocon at the inner envelope membrane of chloroplasts 55-II |
[detail] |
817019 |
| 6.5 |
NADK2 |
NAD kinase 2 |
[detail] |
838766 |
| 6.3 |
AT2G41040 |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein |
[detail] |
818703 |
| 6.1 |
AT1G74640 |
alpha/beta-Hydrolases superfamily protein |
[detail] |
843803 |
| 5.6 |
AT2G40400 |
DUF399 family protein, putative (DUF399 and DUF3411) |
[detail] |
818633 |
|
Coexpressed
gene list |
[Coexpressed gene list for AT1G79600]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
261353_at
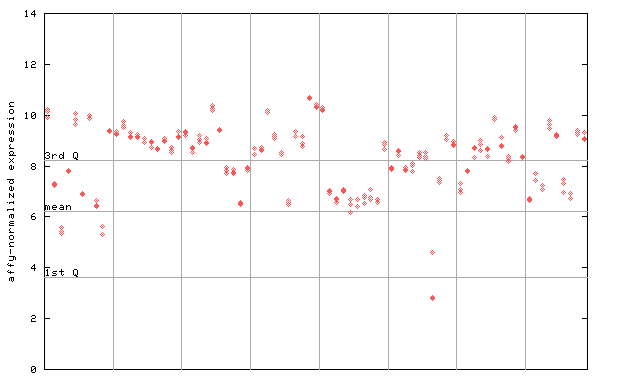
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
261353_at
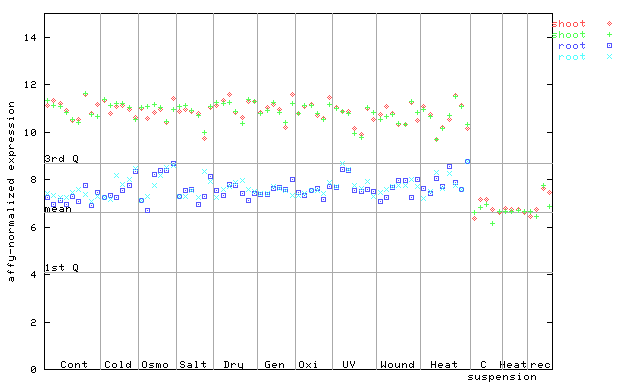
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
261353_at
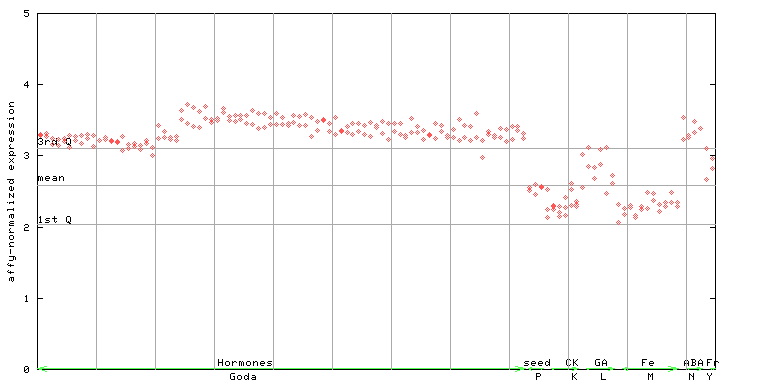
X axis is samples (xls file), and Y axis is log-expression.
|










