| functional annotation |
| Function |
Pyridoxal phosphate (PLP)-dependent transferases superfamily protein |




|
| GO BP |
|
GO:1901997 [list] [network] negative regulation of indoleacetic acid biosynthetic process via tryptophan
|
(1 genes)
|
IDA
|
 |
|
GO:0010366 [list] [network] negative regulation of ethylene biosynthetic process
|
(4 genes)
|
IMP
|
 |
|
GO:0006569 [list] [network] tryptophan catabolic process
|
(5 genes)
|
IMP
|
 |
|
GO:0006570 [list] [network] tyrosine metabolic process
|
(11 genes)
|
IDA
|
 |
|
GO:0006558 [list] [network] L-phenylalanine metabolic process
|
(13 genes)
|
IDA
|
 |
|
GO:0009641 [list] [network] shade avoidance
|
(17 genes)
|
IMP
|
 |
|
GO:0006555 [list] [network] methionine metabolic process
|
(18 genes)
|
IDA
|
 |
|
GO:0009851 [list] [network] auxin biosynthetic process
|
(32 genes)
|
IMP
|
 |
|
GO:0010252 [list] [network] auxin homeostasis
|
(35 genes)
|
IMP
|
 |
|
GO:0006568 [list] [network] tryptophan metabolic process
|
(46 genes)
|
IDA
|
 |
|
GO:0009072 [list] [network] aromatic amino acid metabolic process
|
(91 genes)
|
IDA
|
 |
|
GO:0009698 [list] [network] phenylpropanoid metabolic process
|
(113 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0010326 [list] [network] methionine-oxo-acid transaminase activity
|
(4 genes)
|
IDA
|
 |
|
GO:0008483 [list] [network] transaminase activity
|
(44 genes)
|
IDA
|
 |
|
| KEGG |
ath00270 [list] [network] Cysteine and methionine metabolism (124 genes) |
 |
| ath00380 [list] [network] Tryptophan metabolism (64 genes) |
 |
| Protein |
NP_001322147.1
NP_001322148.1
NP_001322149.1
NP_178152.1
|
| BLAST |
NP_001322147.1
NP_001322148.1
NP_001322149.1
NP_178152.1
|
| Orthologous |
[Ortholog page]
LOC4323866 (osa)
LOC7490978 (ppo)
LOC25491374 (mtr)
LOC100776219 (gma)
LOC100803887 (gma)
LOC101244469 (sly)
LOC103830431 (bra)
LOC103853264 (bra)
LOC123060482 (tae)
LOC123069030 (tae)
LOC123077557 (tae)
LOC123443004 (hvu)
|
Subcellular
localization
wolf |
|
cyto 6,
chlo 2,
pero 1
|
(predict for NP_001322147.1)
|
|
cyto 6,
chlo 2,
pero 1
|
(predict for NP_001322148.1)
|
|
cyto 6,
chlo 2,
pero 1
|
(predict for NP_001322149.1)
|
|
cyto 6,
chlo 2,
pero 1
|
(predict for NP_178152.1)
|
|
Subcellular
localization
TargetP |
|
mito 6,
other 3
|
(predict for NP_001322147.1)
|
|
mito 6,
other 3
|
(predict for NP_001322148.1)
|
|
mito 6,
other 3
|
(predict for NP_001322149.1)
|
|
mito 6,
other 3
|
(predict for NP_178152.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath01230 |
Biosynthesis of amino acids |
11 |
 |
| ath00400 |
Phenylalanine, tyrosine and tryptophan biosynthesis |
6 |
 |
| ath01200 |
Carbon metabolism |
5 |
 |
| ath00710 |
Carbon fixation in photosynthetic organisms |
3 |
 |
| ath00999 |
Biosynthesis of various plant secondary metabolites; Including: Crocin biosynthesis, Cannabidiol biosynthesis, Mugineic acid biosynthesis, Pentagalloylglucose biosynthesis, Benzoxazinoid biosynthesis, Gramine biosynthesis, Coumarin biosynthesis, Furanocoumarin biosynthesis, Hordatine biosynthesis, Podophyllotoxin biosynthesis |
3 |
 |
Genes directly connected with AT1G80360 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 6.3 |
AT5G66120 |
3-dehydroquinate synthase |
[detail] |
836744 |
| 5.7 |
SDRB |
short-chain dehydrogenase-reductase B |
[detail] |
820462 |
| 5.6 |
PDE345 |
Aldolase superfamily protein |
[detail] |
814643 |
|
Coexpressed
gene list |
[Coexpressed gene list for AT1G80360]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
260328_at
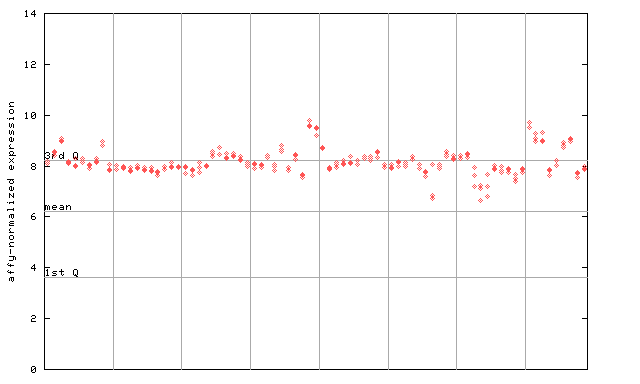
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
260328_at
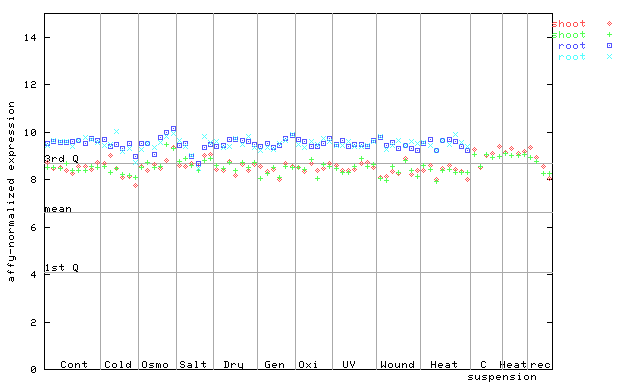
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
260328_at
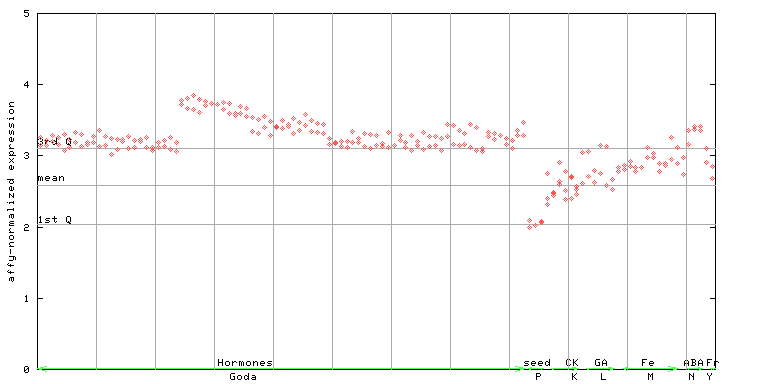
X axis is samples (xls file), and Y axis is log-expression.
|





















