| functional annotation |
| Function |
natural resistance-associated macrophage protein 1 |


|
| GO BP |
|
GO:0070574 [list] [network] cadmium ion transmembrane transport
|
(2 genes)
|
IDA
|
 |
|
GO:0071421 [list] [network] manganese ion transmembrane transport
|
(4 genes)
|
IDA
|
 |
|
GO:0015692 [list] [network] lead ion transport
|
(5 genes)
|
TAS
|
 |
|
GO:0055071 [list] [network] manganese ion homeostasis
|
(6 genes)
|
IMP
|
 |
|
GO:0015691 [list] [network] cadmium ion transport
|
(9 genes)
|
TAS
|
 |
|
GO:0006828 [list] [network] manganese ion transport
|
(11 genes)
|
TAS
|
 |
|
GO:0034755 [list] [network] iron ion transmembrane transport
|
(11 genes)
|
IGI
|
 |
|
GO:0006875 [list] [network] cellular metal ion homeostasis
|
(91 genes)
|
NAS
|
 |
|
GO:0030001 [list] [network] metal ion transport
|
(156 genes)
|
TAS
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0015086 [list] [network] cadmium ion transmembrane transporter activity
|
(7 genes)
|
IDA
|
 |
|
GO:0005384 [list] [network] manganese ion transmembrane transporter activity
|
(14 genes)
|
IDA
IMP
TAS
|
 |
|
GO:0005381 [list] [network] iron ion transmembrane transporter activity
|
(18 genes)
|
IGI
|
 |
|
GO:0015103 [list] [network] inorganic anion transmembrane transporter activity
|
(93 genes)
|
ISS
|
 |
|
GO:0046873 [list] [network] metal ion transmembrane transporter activity
|
(217 genes)
|
TAS
|
 |
|
| KEGG |
|
|
| Protein |
NP_001321606.1
NP_178198.1
|
| BLAST |
NP_001321606.1
NP_178198.1
|
| Orthologous |
[Ortholog page]
NRAMP6 (ath)
LOC9267339 (osa)
LOC18094263 (ppo)
LOC25487976 (mtr)
NRAMP6A (gma)
NRAMP5B (gma)
NRAMP6B (gma)
NRAMP5A (gma)
LOC101246519 (sly)
LOC103830460 (bra)
LOC103832453 (bra)
LOC103852491 (bra)
LOC103853215 (bra)
LOC123166850 (tae)
LOC123412449 (hvu)
|
Subcellular
localization
wolf |
|
chlo 8,
chlo_mito 4,
plas 1,
E.R. 1,
E.R._plas 1
|
(predict for NP_001321606.1)
|
|
chlo 8,
chlo_mito 4,
plas 1,
E.R. 1,
E.R._plas 1
|
(predict for NP_178198.1)
|
|
Subcellular
localization
TargetP |
|
chlo 4
|
(predict for NP_001321606.1)
|
|
chlo 4
|
(predict for NP_178198.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00270 |
Cysteine and methionine metabolism |
2 |
 |
| ath00999 |
Biosynthesis of various plant secondary metabolites; Including: Crocin biosynthesis, Cannabidiol biosynthesis, Mugineic acid biosynthesis, Pentagalloylglucose biosynthesis, Benzoxazinoid biosynthesis, Gramine biosynthesis, Coumarin biosynthesis, Furanocoumarin biosynthesis, Hordatine biosynthesis, Podophyllotoxin biosynthesis |
2 |
 |
Genes directly connected with NRAMP1 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 4.9 |
NIP5;1 |
NOD26-like intrinsic protein 5;1 |
[detail] |
826630 |
| 4.6 |
GTR2 |
Major facilitator superfamily protein |
[detail] |
836389 |
| 4.6 |
MT1C |
metallothionein 1C |
[detail] |
837274 |
|
Coexpressed
gene list |
[Coexpressed gene list for NRAMP1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
261895_at
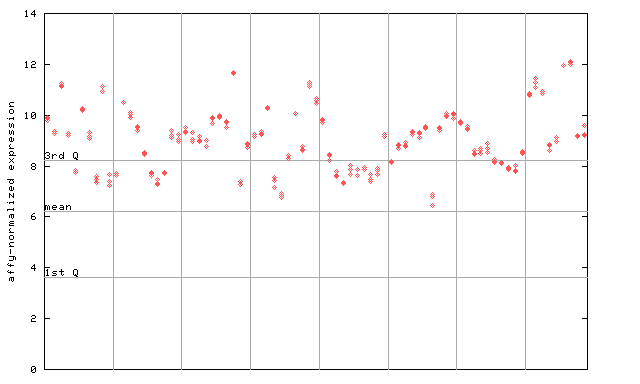
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
261895_at
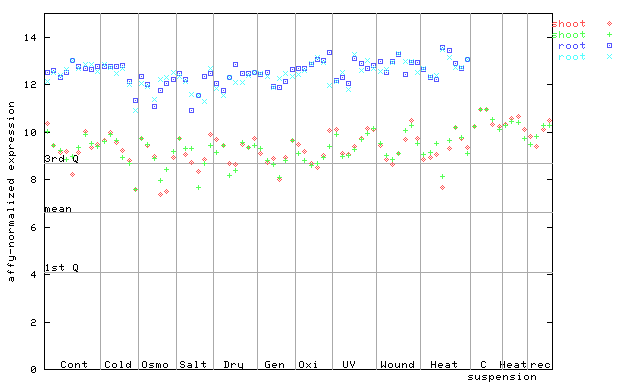
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
261895_at
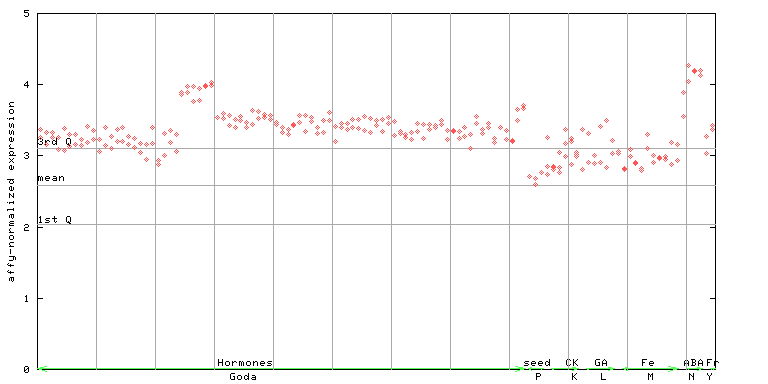
X axis is samples (xls file), and Y axis is log-expression.
|












