| functional annotation |
| Function |
RNI-like superfamily protein |


|
| GO BP |
|
GO:0009641 [list] [network] shade avoidance
|
(17 genes)
|
IMP
|
 |
|
GO:0009861 [list] [network] jasmonic acid and ethylene-dependent systemic resistance
|
(17 genes)
|
TAS
|
 |
|
GO:0009901 [list] [network] anther dehiscence
|
(20 genes)
|
IMP
|
 |
|
GO:0009625 [list] [network] response to insect
|
(32 genes)
|
IMP
|
 |
|
GO:0010218 [list] [network] response to far red light
|
(54 genes)
|
IMP
|
 |
|
GO:0010118 [list] [network] stomatal movement
|
(59 genes)
|
IMP
|
 |
|
GO:0031348 [list] [network] negative regulation of defense response
|
(63 genes)
|
IMP
|
 |
|
GO:0031146 [list] [network] SCF-dependent proteasomal ubiquitin-dependent protein catabolic process
|
(66 genes)
|
IMP
|
 |
|
GO:0009867 [list] [network] jasmonic acid mediated signaling pathway
|
(71 genes)
|
IGI
TAS
|
 |
|
GO:0048443 [list] [network] stamen development
|
(101 genes)
|
IMP
|
 |
|
GO:0009909 [list] [network] regulation of flower development
|
(157 genes)
|
IMP
|
 |
|
GO:0009611 [list] [network] response to wounding
|
(212 genes)
|
IMP
|
 |
|
GO:0009753 [list] [network] response to jasmonic acid
|
(222 genes)
|
IMP
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(399 genes)
|
IMP
|
 |
|
GO:0006511 [list] [network] ubiquitin-dependent protein catabolic process
|
(436 genes)
|
TAS
|
 |
|
GO:0048364 [list] [network] root development
|
(482 genes)
|
IMP
|
 |
|
GO:0050832 [list] [network] defense response to fungus
|
(517 genes)
|
IMP
|
 |
|
GO:0016567 [list] [network] protein ubiquitination
|
(624 genes)
|
IEA
|
 |
|
GO:0006952 [list] [network] defense response
|
(1420 genes)
|
TAS
|
 |
|
| GO CC |
|
GO:0019005 [list] [network] SCF ubiquitin ligase complex
|
(68 genes)
|
IPI
|
 |
|
GO:0005737 [list] [network] cytoplasm
|
(14855 genes)
|
ISM
|
 |
|
| GO MF |
|
| KEGG |
ath04075 [list] [network] Plant hormone signal transduction (273 genes) |
 |
| Protein |
NP_565919.1
|
| BLAST |
NP_565919.1
|
| Orthologous |
[Ortholog page]
Coi1 (sly)
COI1 (gma)
LOC4324818 (osa)
LOC4338955 (osa)
LOC7490347 (ppo)
LOC7494475 (ppo)
LOC11406671 (mtr)
LOC11415803 (mtr)
LOC100254771 (vvi)
LOC100280173 (zma)
LOC100281510 (zma)
LOC100284059 (zma)
LOC100382859 (zma)
LOC100779171 (gma)
LOC100819069 (gma)
LOC103857890 (bra)
LOC103865754 (bra)
LOC103866997 (bra)
|
Subcellular
localization
wolf |
|
cyto 8,
nucl 1,
mito 1,
plas 1,
nucl_plas 1,
mito_plas 1
|
(predict for NP_565919.1)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_565919.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04075 |
Plant hormone signal transduction |
4 |
 |
Genes directly connected with COI1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.5 |
SCL14 |
SCARECROW-like 14 |
[detail] |
837267 |
| 6.3 |
RGA1 |
GRAS family transcription factor family protein |
[detail] |
814686 |
| 6.2 |
AFB3 |
auxin signaling F-box 3 |
[detail] |
837838 |
| 6.0 |
PI4K GAMMA 7 |
phosphoinositide 4-kinase gamma 7 |
[detail] |
814915 |
| 5.8 |
bZIP19 |
Basic-leucine zipper (bZIP) transcription factor family protein |
[detail] |
829656 |
| 5.6 |
AT2G17020 |
F-box/RNI-like superfamily protein |
[detail] |
816205 |
| 5.5 |
OXS2 |
CCCH-type zinc finger protein with ARM repeat domain-containing protein |
[detail] |
818790 |
| 5.2 |
MYC4 |
Basic helix-loop-helix (bHLH) DNA-binding family protein |
[detail] |
827511 |
| 4.7 |
AT2G33360 |
cadherin EGF LAG seven-pass G-type receptor, putative (DUF3527) |
[detail] |
817899 |
| 4.2 |
AT1G01830 |
ARM repeat superfamily protein |
[detail] |
839264 |
|
Coexpressed
gene list |
[Coexpressed gene list for COI1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
267346_at
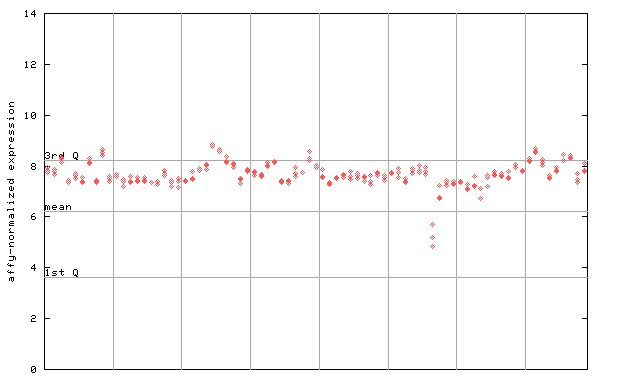
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
267346_at
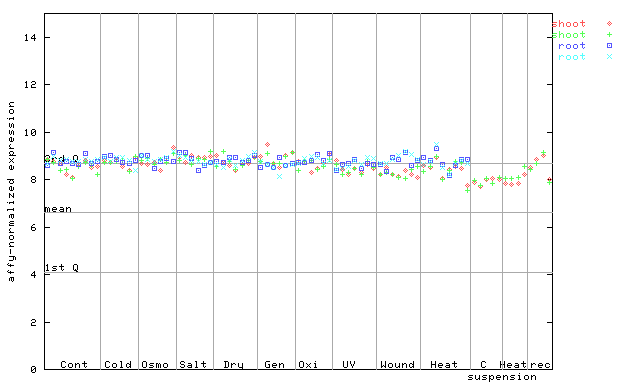
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
267346_at
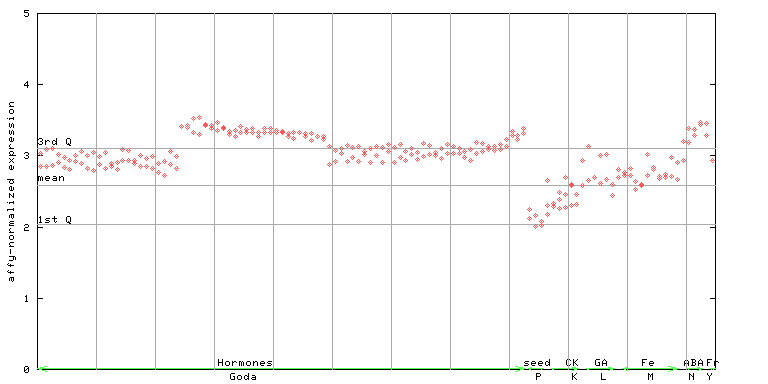
X axis is samples (xls file), and Y axis is log-expression.
|











