| functional annotation |
| Function |
retinoblastoma-related 1 |




|
| GO BP |
|
GO:0022619 [list] [network] generative cell differentiation
|
(1 genes)
|
IMP
|
 |
|
GO:0010377 [list] [network] guard cell fate commitment
|
(2 genes)
|
IGI
|
 |
|
GO:1903866 [list] [network] palisade mesophyll development
|
(2 genes)
|
IMP
|
 |
|
GO:0000082 [list] [network] G1/S transition of mitotic cell cycle
|
(3 genes)
|
IMP
TAS
|
 |
|
GO:2000653 [list] [network] regulation of genetic imprinting
|
(3 genes)
|
IDA
|
 |
|
GO:2000036 [list] [network] regulation of stem cell population maintenance
|
(4 genes)
|
IMP
|
 |
|
GO:0006349 [list] [network] regulation of gene expression by genomic imprinting
|
(15 genes)
|
IPI
|
 |
|
GO:0051783 [list] [network] regulation of nuclear division
|
(19 genes)
|
IMP
|
 |
|
GO:0008356 [list] [network] asymmetric cell division
|
(24 genes)
|
IGI
IMP
|
 |
|
GO:0007129 [list] [network] homologous chromosome pairing at meiosis
|
(29 genes)
|
IMP
|
 |
|
GO:0001708 [list] [network] cell fate specification
|
(34 genes)
|
IMP
|
 |
|
GO:0009960 [list] [network] endosperm development
|
(37 genes)
|
IMP
|
 |
|
GO:0032875 [list] [network] regulation of DNA endoreduplication
|
(38 genes)
|
IMP
|
 |
|
GO:0009567 [list] [network] double fertilization forming a zygote and endosperm
|
(40 genes)
|
IEP
|
 |
|
GO:0010090 [list] [network] trichome morphogenesis
|
(66 genes)
|
IMP
|
 |
|
GO:0001558 [list] [network] regulation of cell growth
|
(132 genes)
|
IMP
|
 |
|
GO:0051726 [list] [network] regulation of cell cycle
|
(158 genes)
|
IMP
|
 |
|
GO:0009553 [list] [network] embryo sac development
|
(163 genes)
|
IMP
|
 |
|
GO:0009555 [list] [network] pollen development
|
(376 genes)
|
IMP
|
 |
|
GO:0048366 [list] [network] leaf development
|
(465 genes)
|
IMP
|
 |
|
GO:0048229 [list] [network] gametophyte development
|
(523 genes)
|
IMP
|
 |
|
GO:0030154 [list] [network] cell differentiation
|
(1176 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001189868.1
NP_566417.3
|
| BLAST |
NP_001189868.1
NP_566417.3
|
| Orthologous |
[Ortholog page]
LOC4346176 (osa)
LOC7455806 (ppo)
LOC11429529 (mtr)
LOC100137000 (tae)
LOC100777429 (gma)
LOC100777805 (gma)
LOC100779266 (gma)
RBR1 (sly)
LOC103249150 (bra)
LOC103859438 (bra)
LOC103870260 (bra)
LOC123152064 (tae)
LOC123165294 (tae)
LOC123407429 (hvu)
|
Subcellular
localization
wolf |
|
nucl 4,
cyto 2,
nucl_plas 2,
E.R. 1,
cyto_plas 1
|
(predict for NP_001189868.1)
|
|
nucl 4,
cyto 2,
nucl_plas 2,
E.R. 1,
cyto_plas 1
|
(predict for NP_566417.3)
|
|
Subcellular
localization
TargetP |
|
other 8,
chlo 3
|
(predict for NP_001189868.1)
|
|
other 8,
chlo 3
|
(predict for NP_566417.3)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03030 |
DNA replication |
10 |
 |
Genes directly connected with RBR1 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 7.9 |
RNR1 |
ribonucleotide reductase 1 |
[detail] |
816715 |
| 7.9 |
POLA2 |
DNA polymerase alpha 2 |
[detail] |
843086 |
| 7.4 |
MCM5 |
Minichromosome maintenance (MCM2/3/5) family protein |
[detail] |
815415 |
| 7.1 |
AT3G21480 |
BRCT domain-containing DNA repair protein |
[detail] |
821702 |
|
Coexpressed
gene list |
[Coexpressed gene list for RBR1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
256268_at
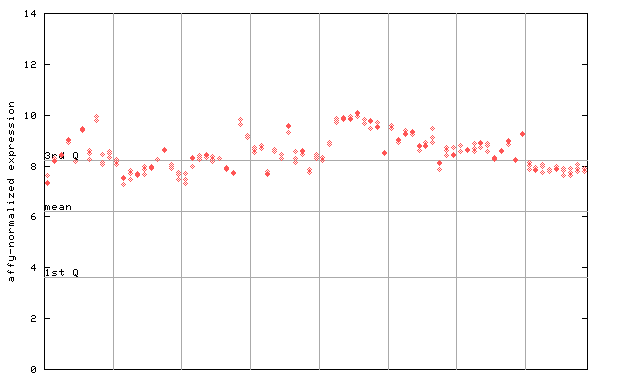
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
256268_at
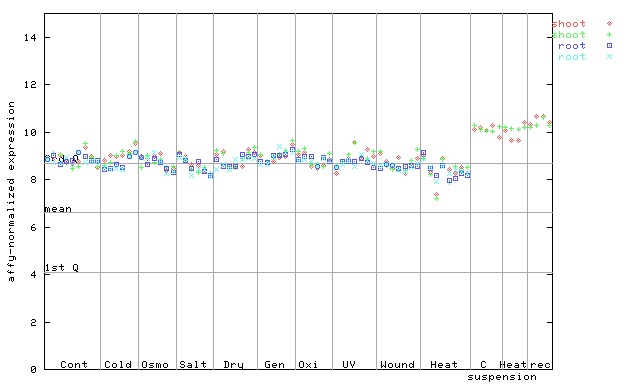
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
256268_at
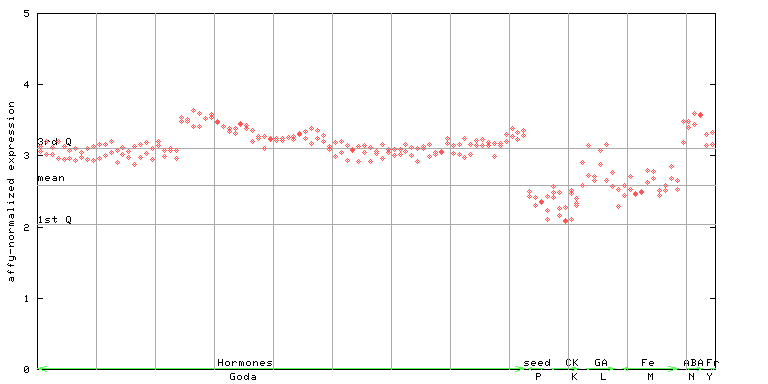
X axis is samples (xls file), and Y axis is log-expression.
|













