[←][→] ath
| functional annotation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | 2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |




|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | ath00999 [list] [network] Biosynthesis of various plant secondary metabolites; Including: Crocin biosynthesis, Cannabidiol biosynthesis, Mugineic acid biosynthesis, Pentagalloylglucose biosynthesis, Benzoxazinoid biosynthesis, Gramine biosynthesis, Coumarin biosynthesis, Furanocoumarin biosynthesis, Hordatine biosynthesis, Podophyllotoxin biosynthesis (66 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_187896.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_187896.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] LOC7480442 (ppo) LOC11407206 (mtr) LOC11409249 (mtr) LOC100803722 (gma) LOC101262174 (sly) LOC102659647 (gma) LOC103849720 (bra) LOC103859475 (bra) LOC103870212 (bra) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for AT3G12900] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
257135_at
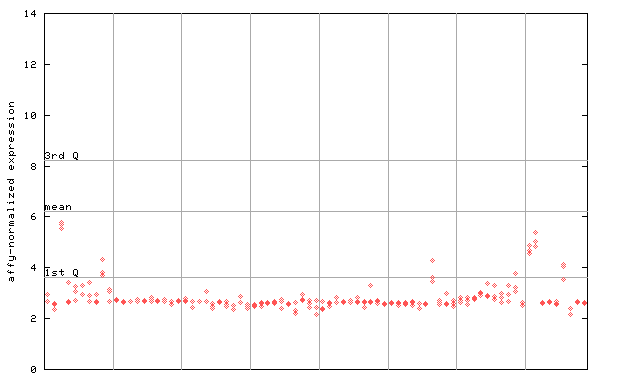
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
257135_at
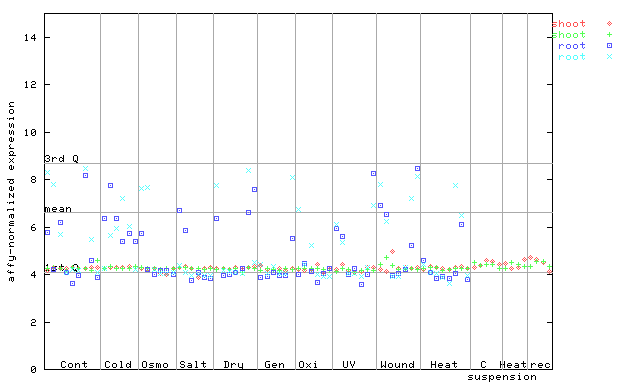
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
257135_at
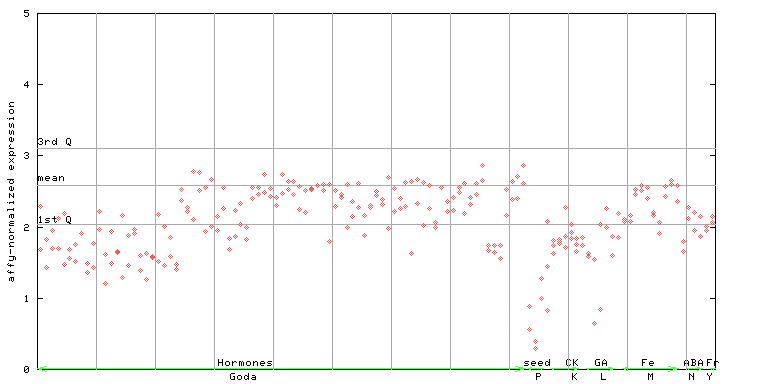
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 820473 |


|
| Refseq ID (protein) | NP_187896.1 |  |
The preparation time of this page was 0.2 [sec].



