| functional annotation |
| Function |
Nucleotide-diphospho-sugar transferases superfamily protein |


|
| GO BP |
|
GO:0010401 [list] [network] pectic galactan metabolic process
|
(1 genes)
|
IMP
|
 |
|
GO:0052541 [list] [network] plant-type cell wall cellulose metabolic process
|
(13 genes)
|
IMP
|
 |
|
GO:0007155 [list] [network] cell adhesion
|
(14 genes)
|
IEA
|
 |
|
GO:0006665 [list] [network] sphingolipid metabolic process
|
(45 genes)
|
IMP
|
 |
|
GO:0010087 [list] [network] phloem or xylem histogenesis
|
(102 genes)
|
IMP
|
 |
|
GO:0008219 [list] [network] cell death
|
(127 genes)
|
IMP
|
 |
|
GO:0006486 [list] [network] protein glycosylation
|
(134 genes)
|
IEA
|
 |
|
GO:0009738 [list] [network] abscisic acid-activated signaling pathway
|
(194 genes)
|
IEA
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(574 genes)
|
IMP
|
 |
|
GO:0007275 [list] [network] multicellular organism development
|
(2714 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0035251 [list] [network] UDP-glucosyltransferase activity
|
(141 genes)
|
ISS
|
 |
|
GO:0016757 [list] [network] transferase activity, transferring glycosyl groups
|
(644 genes)
|
IMP
ISS
|
 |
|
GO:0046872 [list] [network] metal ion binding
|
(3180 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_191142.1
|
| BLAST |
NP_191142.1
|
| Orthologous |
[Ortholog page]
LOC7488010 (ppo)
LOC7490339 (ppo)
LOC9267681 (osa)
LOC11426546 (mtr)
LOC100260303 (vvi)
LOC100776313 (gma)
LOC100781217 (gma)
LOC100781422 (gma)
LOC100797004 (gma)
LOC103626422 (zma)
LOC103841490 (bra)
|
Subcellular
localization
wolf |
|
cyto 5,
nucl 1,
chlo 1,
mito 1,
extr 1,
E.R. 1,
cysk 1,
chlo_mito 1,
nucl_plas 1
|
(predict for NP_191142.1)
|
|
Subcellular
localization
TargetP |
|
other 9
|
(predict for NP_191142.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04016 |
MAPK signaling pathway - plant |
2 |
 |
Genes directly connected with EPC1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 6.9 |
AT4G31170 |
Protein kinase superfamily protein |
[detail] |
829245 |
| 6.8 |
MNS3 |
alpha-mannosidase 3 |
[detail] |
839879 |
| 6.6 |
AT2G36360 |
Galactose oxidase/kelch repeat superfamily protein |
[detail] |
818209 |
| 6.1 |
AT5G43100 |
Eukaryotic aspartyl protease family protein |
[detail] |
834326 |
| 6.0 |
MPK2 |
mitogen-activated protein kinase homolog 2 |
[detail] |
842248 |
| 6.0 |
AT1G13000 |
transmembrane protein, putative (DUF707) |
[detail] |
837858 |
| 5.6 |
AT5G42000 |
ORMDL family protein |
[detail] |
834205 |
| 5.4 |
RGS1 |
REGULATOR OF G-PROTEIN SIGNALING 1 |
[detail] |
822207 |
|
Coexpressed
gene list |
[Coexpressed gene list for EPC1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
251764_at
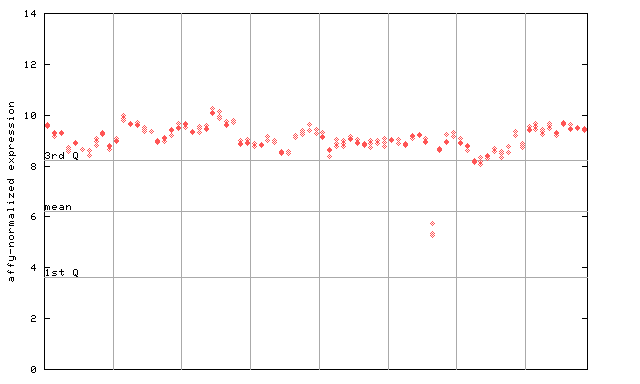
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
251764_at
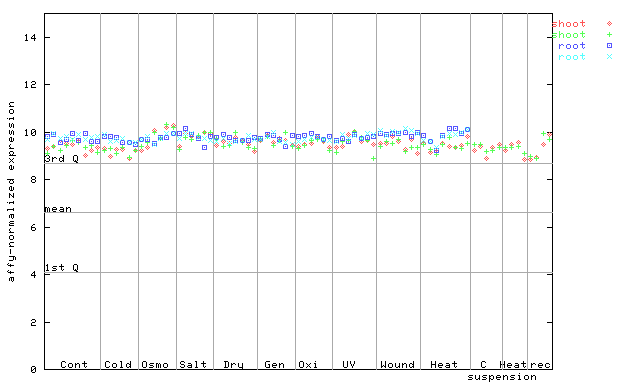
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
251764_at
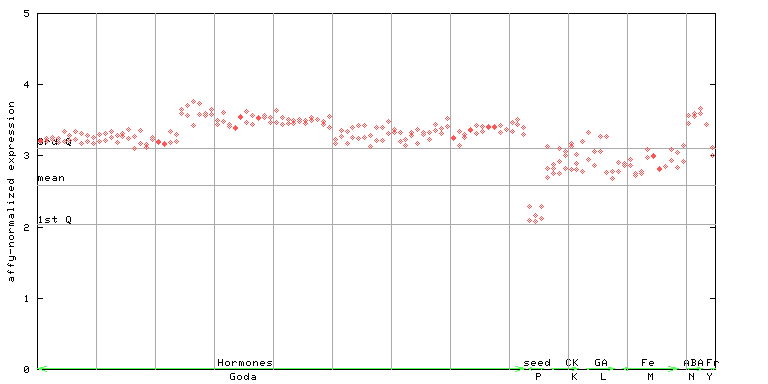
X axis is samples (xls file), and Y axis is log-expression.
|










