[←][→] ath
| functional annotation | ||||||||||
| Function | citrate synthase 5 |


|
||||||||
| GO BP |
|
|||||||||
| GO CC |
|
|||||||||
| GO MF |
|
|||||||||
| KEGG | ath00020 [list] [network] Citrate cycle (TCA cycle) (64 genes) |  |
||||||||
| ath00630 [list] [network] Glyoxylate and dicarboxylate metabolism (78 genes) |  |
|||||||||
| ath01200 [list] [network] Carbon metabolism (273 genes) |  |
|||||||||
| ath01210 [list] [network] 2-Oxocarboxylic acid metabolism (74 genes) |  |
|||||||||
| ath01230 [list] [network] Biosynthesis of amino acids (244 genes) |  |
|||||||||
| Protein | NP_001327512.1 NP_001327513.1 NP_191569.2 | |||||||||
| BLAST | NP_001327512.1 NP_001327513.1 NP_191569.2 | |||||||||
| Orthologous | [Ortholog page] ATCS (ath) LOC4328598 (osa) LOC4350660 (osa) LOC18094999 (ppo) LOC25486862 (mtr) LOC100499624 (gma) LOC100781418 (gma) LOC101249011 (sly) LOC103858130 (bra) LOC103862913 (bra) LOC103866133 (bra) LOC123132662 (tae) LOC123136754 (tae) LOC123144240 (tae) LOC123401217 (hvu) | |||||||||
| Subcellular localization wolf |
|
|||||||||
| Subcellular localization TargetP |
|
|||||||||
| Gene coexpression | ||||||||||
| Network*for coexpressed genes |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for CSY5] | |||||||||
| Gene expression | ||||||||||
| All samples | [Expression pattern for all samples] | |||||||||
| AtGenExpress* (Development) |
251455_at
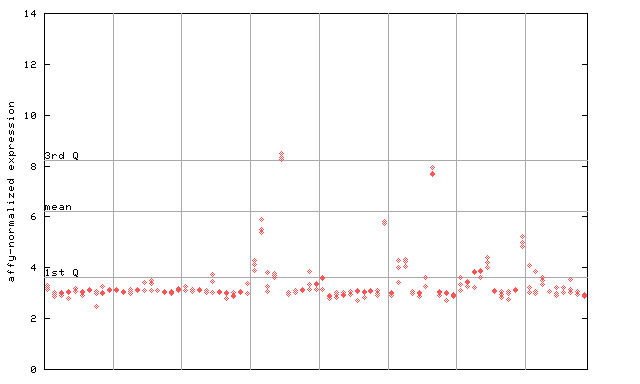
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||
| AtGenExpress* (Stress) |
251455_at
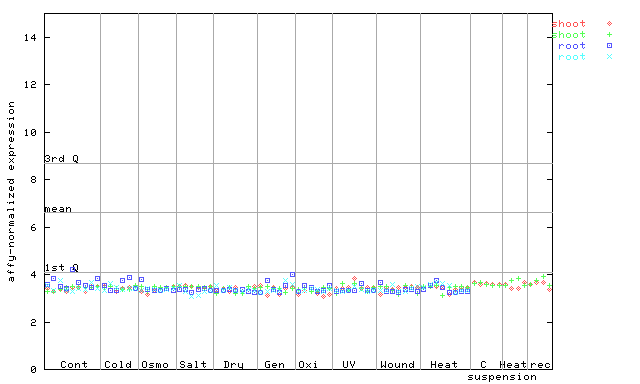
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||
| AtGenExpress* (Hormone) |
251455_at
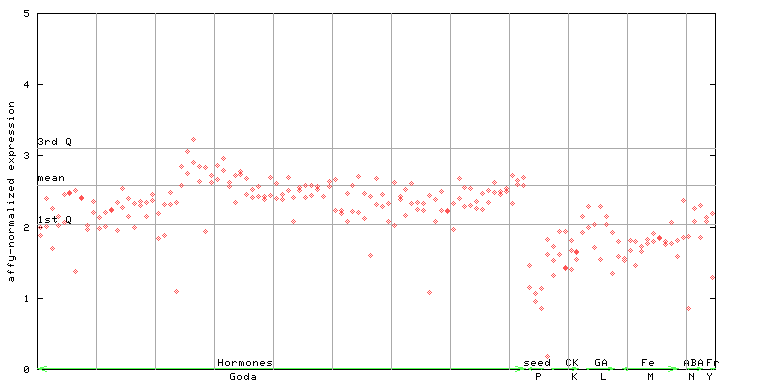
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||
| Link to other DBs | ||
| Entrez Gene ID | 825180 |


|
| Refseq ID (protein) | NP_001327512.1 |  |
| NP_001327513.1 |  |
|
| NP_191569.2 |  |
|
The preparation time of this page was 0.1 [sec].

