| functional annotation |
| Function |
RNA polymerase II transcription mediator |


|
| GO BP |
|
GO:0090213 [list] [network] regulation of radial pattern formation
|
(2 genes)
|
IMP
|
 |
|
GO:0040034 [list] [network] regulation of development, heterochronic
|
(49 genes)
|
IMP
|
 |
|
GO:0048573 [list] [network] photoperiodism, flowering
|
(67 genes)
|
NAS
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_001319832.1
NP_001329123.1
NP_001329124.1
|
| BLAST |
NP_001319832.1
NP_001329123.1
NP_001329124.1
|
| Orthologous |
[Ortholog page]
LOC4344116 (osa)
LOC4349313 (osa)
LOC7494075 (ppo)
LOC11442155 (mtr)
LOC11444032 (mtr)
LOC100263628 (vvi)
LOC100384108 (zma)
LOC100781696 (gma)
LOC100782672 (gma)
LOC100806985 (gma)
LOC100811040 (gma)
LOC101246089 (sly)
LOC103630556 (zma)
LOC103836919 (bra)
LOC103858870 (bra)
|
Subcellular
localization
wolf |
|
chlo 4,
nucl 4,
E.R. 1,
plas 1,
pero 1,
E.R._vacu 1,
cyto_E.R. 1
|
(predict for NP_001319832.1)
|
|
chlo 4,
nucl 4,
E.R. 1,
plas 1,
pero 1,
E.R._vacu 1,
cyto_E.R. 1
|
(predict for NP_001329123.1)
|
|
chlo 4,
nucl 4,
E.R. 1,
plas 1,
pero 1,
E.R._vacu 1,
cyto_E.R. 1
|
(predict for NP_001329124.1)
|
|
Subcellular
localization
TargetP |
|
chlo 7
|
(predict for NP_001319832.1)
|
|
chlo 7
|
(predict for NP_001329123.1)
|
|
chlo 7
|
(predict for NP_001329124.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with CCT on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 14.3 |
KAK |
HECT ubiquitin protein ligase family protein KAK |
[detail] |
830017 |
| 14.3 |
AT3G52250 |
Duplicated homeodomain-like superfamily protein |
[detail] |
824390 |
| 14.1 |
AT1G02080 |
transcription regulator |
[detail] |
839244 |
| 13.3 |
AT3G50370 |
hypothetical protein |
[detail] |
824199 |
| 12.2 |
AT5G15680 |
ARM repeat superfamily protein |
[detail] |
831422 |
| 11.7 |
RAPTOR1 |
Regulatory-associated protein of TOR 1 |
[detail] |
820032 |
| 10.6 |
AT1G72390 |
nuclear receptor coactivator |
[detail] |
843571 |
| 10.5 |
SUO |
protein SUO |
[detail] |
823960 |
| 10.4 |
MBD9 |
methyl-CPG-binding domain 9 |
[detail] |
821132 |
| 10.3 |
SWP |
RNA polymerase II transcription mediator |
[detail] |
819634 |
| 10.1 |
AT3G11960 |
Cleavage and polyadenylation specificity factor (CPSF) A subunit protein |
[detail] |
820369 |
| 9.7 |
AT5G07940 |
dentin sialophosphoprotein-like protein |
[detail] |
830688 |
| 9.4 |
FAB1A |
1-phosphatidylinositol-3-phosphate 5-kinase FAB1A |
[detail] |
829460 |
| 9.4 |
AT3G61690 |
nucleotidyltransferase |
[detail] |
825342 |
| 9.1 |
AT3G48060 |
BAH and TFIIS domain-containing protein |
[detail] |
823961 |
| 8.8 |
TIC |
time for coffee |
[detail] |
821807 |
| 8.6 |
AT3G51120 |
zinc finger CCCH domain-containing protein 44 |
[detail] |
824276 |
| 7.9 |
AT3G24880 |
Helicase/SANT-associated, DNA binding protein |
[detail] |
822086 |
| 7.7 |
SLOMO |
F-box family protein |
[detail] |
829457 |
| 7.3 |
FRS8 |
FAR1-related sequence 8 |
[detail] |
844341 |
| 7.2 |
GSL1 |
glucan synthase-like 1 |
[detail] |
825838 |
| 6.9 |
AT5G19420 |
Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain-containing protein |
[detail] |
832062 |
| 6.6 |
TPL |
Transducin family protein / WD-40 repeat family protein |
[detail] |
838144 |
|
Coexpressed
gene list |
[Coexpressed gene list for CCT]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
255665_at
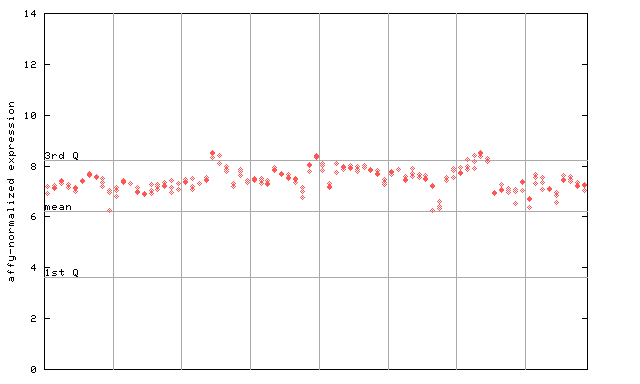
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
255665_at
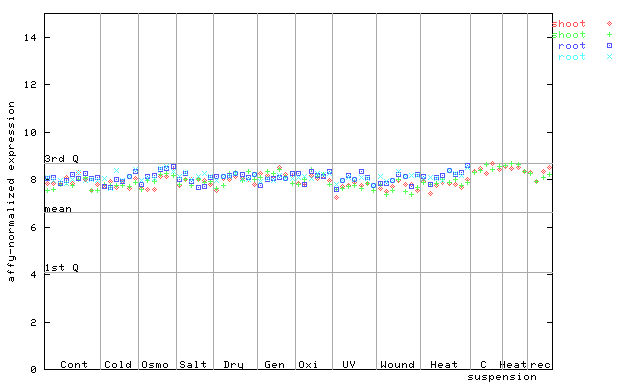
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
255665_at
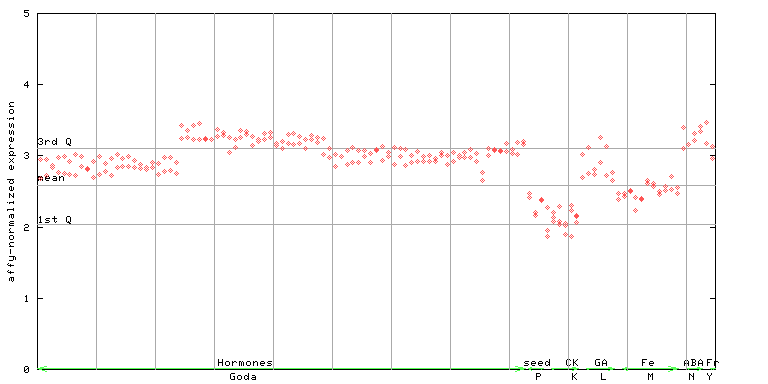
X axis is samples (xls file), and Y axis is log-expression.
|











