| functional annotation |
| Function |
MutT/nudix family protein |

 Plant GARDEN Plant GARDEN JBrowse
Plant GARDEN Plant GARDEN JBrowse
|
| GO BP |
|
GO:0070212 [list] [network] protein poly-ADP-ribosylation
|
(3 genes)
|
IDA
|
 |
|
GO:0010581 [list] [network] regulation of starch biosynthetic process
|
(8 genes)
|
IMP
|
 |
|
GO:0010193 [list] [network] response to ozone
|
(36 genes)
|
IEP
|
 |
|
GO:0002758 [list] [network] innate immune response-activating signaling pathway
|
(38 genes)
|
IMP
|
 |
|
GO:0009626 [list] [network] plant-type hypersensitive response
|
(46 genes)
|
IMP
|
 |
|
GO:0071456 [list] [network] cellular response to hypoxia
|
(241 genes)
|
HEP
|
 |
|
GO:0006979 [list] [network] response to oxidative stress
|
(545 genes)
|
IEP
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(607 genes)
|
IEP
|
 |
|
GO:0042742 [list] [network] defense response to bacterium
|
(980 genes)
|
IMP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0080042 [list] [network] ADP-glucose pyrophosphohydrolase activity
|
(2 genes)
|
IDA
|
 |
|
GO:0017110 [list] [network] nucleoside diphosphate phosphatase activity
|
(3 genes)
|
IDA
|
 |
|
GO:0080041 [list] [network] ADP-ribose pyrophosphohydrolase activity
|
(3 genes)
|
IDA
|
 |
|
GO:0000210 [list] [network] NAD+ diphosphatase activity
|
(5 genes)
|
IDA
|
 |
|
GO:0047631 [list] [network] ADP-ribose diphosphatase activity
|
(6 genes)
|
IDA
|
 |
|
GO:0051287 [list] [network] NAD binding
|
(29 genes)
|
IDA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(5066 genes)
|
IPI
|
 |
|
| KEGG |
|
|
| Protein |
NP_001190707.1
NP_001329573.1
NP_001329574.1
NP_193008.1
NP_849367.1
NP_849368.1
|
| BLAST |
NP_001190707.1
NP_001329573.1
NP_001329574.1
NP_193008.1
NP_849367.1
NP_849368.1
|
| Orthologous |
[Ortholog page]
NUDT5 (ath)
NUDT6 (ath)
LOC103833538 (bra)
LOC103860821 (bra)
|
Subcellular
localization
wolf |
|
cyto 6,
nucl 1,
cysk 1,
cysk_nucl 1
|
(predict for NP_001190707.1)
|
|
cyto 4,
nucl 4,
chlo 1
|
(predict for NP_001329573.1)
|
|
cyto 5,
pero 2,
chlo 1,
extr 1,
cysk 1,
golg 1
|
(predict for NP_001329574.1)
|
|
cysk 4,
cyto 4,
nucl 1
|
(predict for NP_193008.1)
|
|
cysk 4,
cyto 4,
nucl 1
|
(predict for NP_849367.1)
|
|
cysk 4,
cyto 4,
nucl 1
|
(predict for NP_849368.1)
|
|
Subcellular
localization
TargetP |
|
other 8
|
(predict for NP_001190707.1)
|
|
other 7
|
(predict for NP_001329573.1)
|
|
other 8
|
(predict for NP_001329574.1)
|
|
other 8
|
(predict for NP_193008.1)
|
|
other 8
|
(predict for NP_849367.1)
|
|
other 8
|
(predict for NP_849368.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04626 |
Plant-pathogen interaction |
5 |
 |
Genes directly connected with NUDT7 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 13.1 |
TCH3 |
Calcium-binding EF hand family protein |
[detail] |
818709 |
| 12.4 |
GILP |
GSH-induced LITAF domain protein |
[detail] |
831158 |
| 10.8 |
ACA1 |
autoinhibited Ca2+-ATPase 1 |
[detail] |
839670 |
| 10.5 |
SYP121 |
syntaxin of plants 121 |
[detail] |
820355 |
| 9.8 |
CPK28 |
calcium-dependent protein kinase 28 |
[detail] |
836753 |
| 9.7 |
AT4G34150 |
Calcium-dependent lipid-binding (CaLB domain) family protein |
[detail] |
829563 |
| 9.3 |
AT2G20142 |
Toll-Interleukin-Resistance (TIR) domain family protein |
[detail] |
2745551 |
| 9.0 |
BCS1 |
cytochrome BC1 synthesi |
[detail] |
824257 |
| 9.0 |
AT1G69840 |
SPFH/Band 7/PHB domain-containing membrane-associated protein family |
[detail] |
843320 |
| 7.9 |
CPK4 |
calcium-dependent protein kinase 4 |
[detail] |
826541 |
| 7.7 |
CNGC19 |
cyclic nucleotide gated channel 19 |
[detail] |
821036 |
| 7.2 |
S6K2 |
serine/threonine protein kinase 2 |
[detail] |
820019 |
|
Coexpressed
gene list |
[Coexpressed gene list for NUDT7]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
254784_at
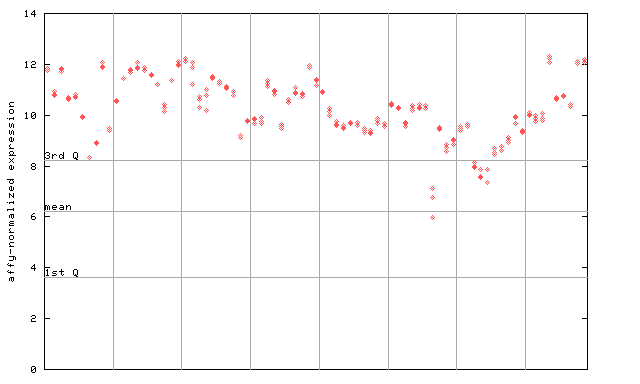
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
254784_at
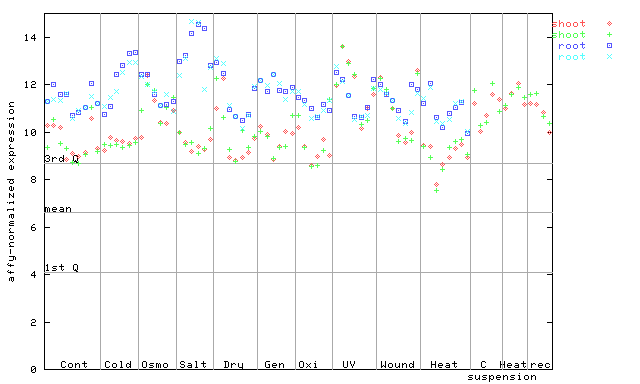
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
254784_at
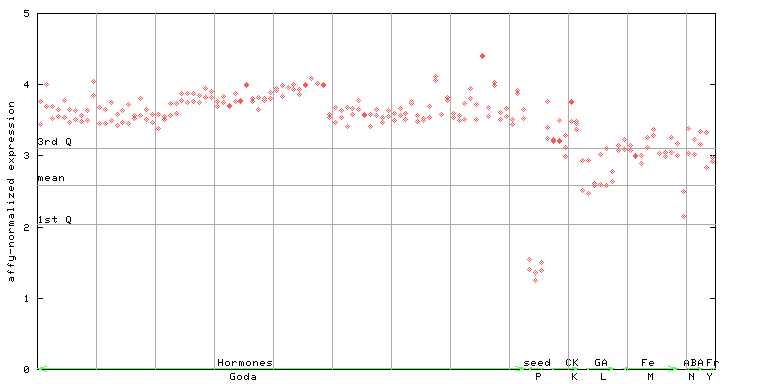
X axis is samples (xls file), and Y axis is log-expression.
|

 Plant GARDEN Plant GARDEN JBrowse
Plant GARDEN Plant GARDEN JBrowse













