| functional annotation |
| Function |
Transducin/WD40 repeat-like superfamily protein |




|
| GO BP |
|
GO:0044770 [list] [network] cell cycle phase transition
|
(10 genes)
|
IEA
|
 |
|
GO:0061640 [list] [network] cytoskeleton-dependent cytokinesis
|
(43 genes)
|
IEA
|
 |
|
GO:0006261 [list] [network] DNA-templated DNA replication
|
(44 genes)
|
IEA
|
 |
|
GO:0000911 [list] [network] cytokinesis by cell plate formation
|
(70 genes)
|
IEA
|
 |
|
GO:0000278 [list] [network] mitotic cell cycle
|
(86 genes)
|
IEA
|
 |
|
GO:0140013 [list] [network] meiotic nuclear division
|
(91 genes)
|
IEA
|
 |
|
GO:0010564 [list] [network] regulation of cell cycle process
|
(92 genes)
|
IEA
|
 |
|
GO:0033043 [list] [network] regulation of organelle organization
|
(94 genes)
|
IEA
|
 |
|
GO:0009658 [list] [network] chloroplast organization
|
(209 genes)
|
IEA
|
 |
|
GO:0051301 [list] [network] cell division
|
(442 genes)
|
IEA
|
 |
|
GO:0048229 [list] [network] gametophyte development
|
(523 genes)
|
IEA
|
 |
|
GO:0022607 [list] [network] cellular component assembly
|
(625 genes)
|
IEA
|
 |
|
GO:0009793 [list] [network] embryo development ending in seed dormancy
|
(697 genes)
|
IEA
|
 |
|
GO:0071554 [list] [network] cell wall organization or biogenesis
|
(898 genes)
|
IEA
|
 |
|
GO:0048523 [list] [network] negative regulation of cellular process
|
(960 genes)
|
IEA
|
 |
|
GO:0009888 [list] [network] tissue development
|
(1806 genes)
|
IEA
|
 |
|
| GO CC |
|
| GO MF |
|
| KEGG |
|
|
| Protein |
NP_193167.3
NP_849378.2
|
| BLAST |
NP_193167.3
NP_849378.2
|
| Orthologous |
[Ortholog page]
LOC4331091 (osa)
LOC7459974 (ppo)
LOC7473467 (ppo)
LOC25482986 (mtr)
LOC100785231 (gma)
LOC100809470 (gma)
LOC101263160 (sly)
LOC103835201 (bra)
LOC123128917 (tae)
LOC123139518 (tae)
LOC123146074 (tae)
LOC123404330 (hvu)
|
Subcellular
localization
wolf |
|
nucl 5,
chlo 3,
cyto_nucl 3
|
(predict for NP_193167.3)
|
|
nucl 4,
chlo 3,
mito 1,
cyto 1,
cyto_mito 1,
golg_plas 1
|
(predict for NP_849378.2)
|
|
Subcellular
localization
TargetP |
|
chlo 5
|
(predict for NP_193167.3)
|
|
chlo 5
|
(predict for NP_849378.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00270 |
Cysteine and methionine metabolism |
2 |
 |
| ath03030 |
DNA replication |
2 |
 |
Genes directly connected with AT4G14310 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 11.8 |
AT2G36200 |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
818192 |
| 11.0 |
AT3G54750 |
downstream neighbor of Son |
[detail] |
824640 |
| 11.0 |
MET1 |
methyltransferase 1 |
[detail] |
834975 |
| 10.7 |
VIM1 |
Zinc finger (C3HC4-type RING finger) family protein |
[detail] |
842157 |
| 9.1 |
EMB2656 |
ARM repeat superfamily protein |
[detail] |
833741 |
| 9.0 |
CMT3 |
chromomethylase 3 |
[detail] |
843313 |
|
Coexpressed
gene list |
[Coexpressed gene list for AT4G14310]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
245606_at
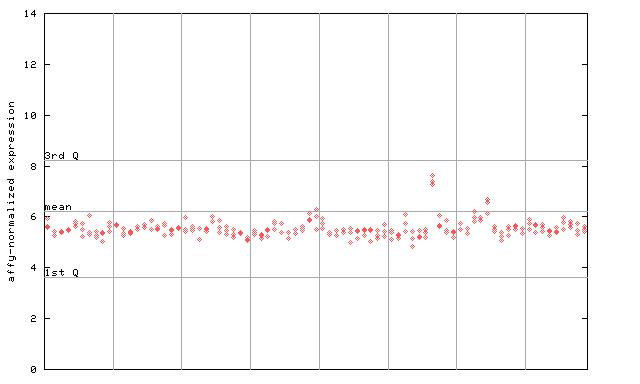
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
245606_at
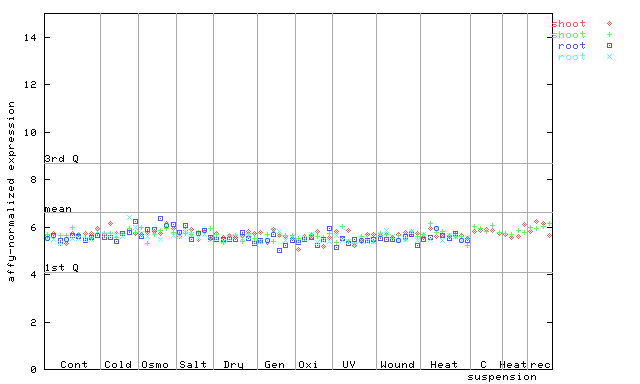
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
245606_at
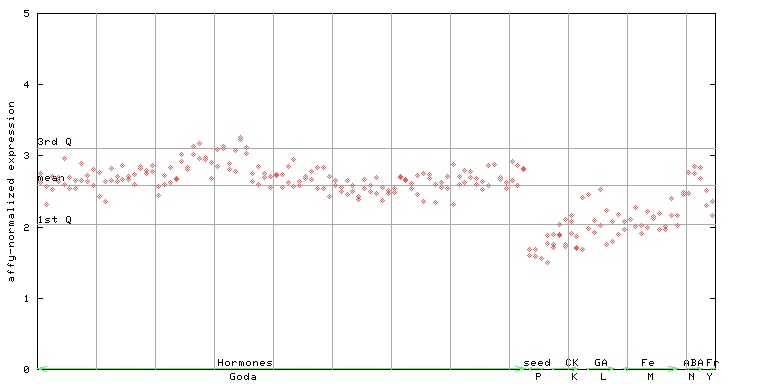
X axis is samples (xls file), and Y axis is log-expression.
|














