[←][→] ath
| functional annotation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | cyclin-dependent kinase-activating kinase assembly factor-related / CDK-activating kinase assembly factor-like protein |




|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | ath03022 [list] [network] Basal transcription factors (55 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath03420 [list] [network] Nucleotide excision repair (62 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_001078472.1 NP_001328131.1 NP_001328132.1 NP_001328133.1 NP_001328134.1 NP_001328135.1 NP_001328136.1 NP_001328137.1 NP_001328138.1 NP_001328139.1 NP_001328140.1 NP_001328141.1 NP_001328142.1 NP_001328143.1 NP_001328144.1 NP_001328145.1 NP_001328146.1 NP_001328147.1 NP_001328148.1 NP_001328149.1 NP_001328150.1 NP_001328151.1 NP_001328152.1 NP_001328153.1 NP_001328154.1 NP_567855.1 NP_849476.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_001078472.1 NP_001328131.1 NP_001328132.1 NP_001328133.1 NP_001328134.1 NP_001328135.1 NP_001328136.1 NP_001328137.1 NP_001328138.1 NP_001328139.1 NP_001328140.1 NP_001328141.1 NP_001328142.1 NP_001328143.1 NP_001328144.1 NP_001328145.1 NP_001328146.1 NP_001328147.1 NP_001328148.1 NP_001328149.1 NP_001328150.1 NP_001328151.1 NP_001328152.1 NP_001328153.1 NP_001328154.1 NP_567855.1 NP_849476.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] LOC4340268 (osa) LOC4350497 (osa) LOC7485263 (ppo) LOC7489425 (ppo) LOC11434892 (mtr) LOC100305654 (gma) LOC101265536 (sly) LOC103834652 (bra) LOC103862006 (bra) LOC123152550 (tae) LOC123160923 (tae) LOC123167001 (tae) LOC123410832 (hvu) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for AT4G30820] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
253594_at
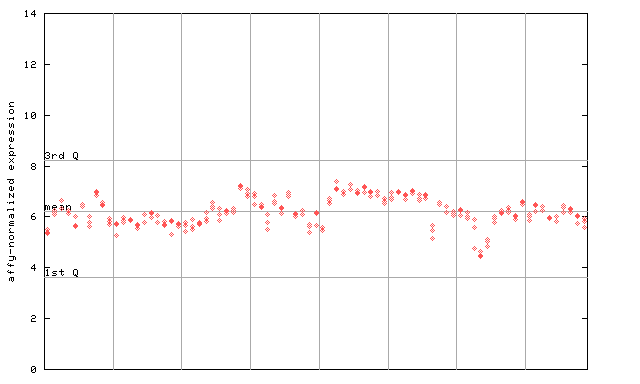
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
253594_at
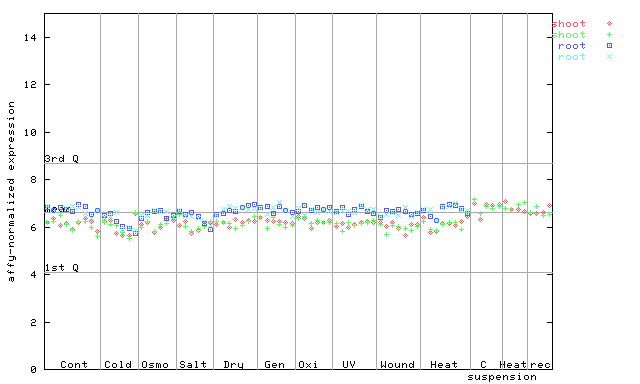
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
253594_at
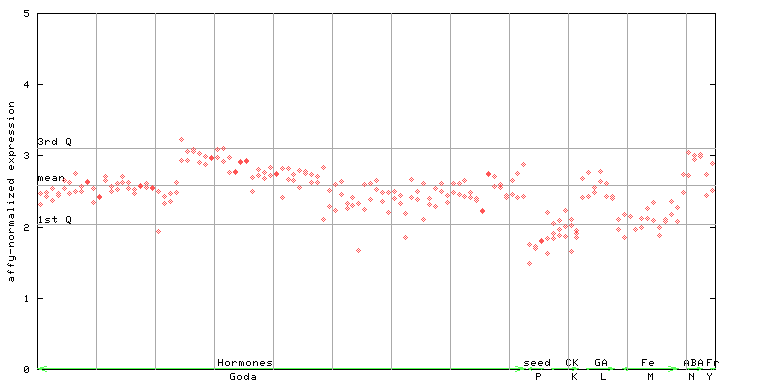
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
The preparation time of this page was 0.2 [sec].


