| functional annotation |
| Function |
plant glycogenin-like starch initiation protein 3 |


|
| GO BP |
|
GO:0010417 [list] [network] glucuronoxylan biosynthetic process
|
(12 genes)
|
IMP
|
 |
|
GO:0045492 [list] [network] xylan biosynthetic process
|
(35 genes)
|
IBA
IDA
IGI
|
 |
|
GO:0009834 [list] [network] plant-type secondary cell wall biogenesis
|
(58 genes)
|
IGI
|
 |
|
GO:0071555 [list] [network] cell wall organization
|
(538 genes)
|
IEA
|
 |
|
| GO CC |
|
GO:0000139 [list] [network] Golgi membrane
|
(404 genes)
|
IEA
|
 |
|
GO:0005794 [list] [network] Golgi apparatus
|
(1430 genes)
|
IDA
ISM
|
 |
|
GO:0016021 [list] [network] integral component of membrane
|
(4803 genes)
|
IEA
|
 |
|
| GO MF |
|
GO:0080116 [list] [network] glucuronoxylan glucuronosyltransferase activity
|
(4 genes)
|
IMP
|
 |
|
GO:0015020 [list] [network] glucuronosyltransferase activity
|
(21 genes)
|
IDA
|
 |
|
GO:0016757 [list] [network] transferase activity, transferring glycosyl groups
|
(644 genes)
|
ISS
|
 |
|
GO:0046872 [list] [network] metal ion binding
|
(3180 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001154284.1
NP_001328034.1
NP_001328035.1
NP_001328036.1
NP_195059.3
|
| BLAST |
NP_001154284.1
NP_001328034.1
NP_001328035.1
NP_001328036.1
NP_195059.3
|
| Orthologous |
[Ortholog page]
LOC4331856 (osa)
LOC11412568 (mtr)
LOC100191941 (zma)
LOC100266116 (vvi)
LOC100780684 (gma)
LOC100795248 (gma)
LOC100812365 (gma)
LOC101255391 (sly)
LOC101260637 (sly)
LOC103834490 (bra)
LOC103848105 (bra)
|
Subcellular
localization
wolf |
|
chlo 6,
chlo_mito 4,
cyto 2,
plas 1,
extr 1
|
(predict for NP_001154284.1)
|
|
cyto 6,
nucl 1,
E.R. 1,
chlo 1,
vacu 1,
cysk_nucl 1,
nucl_plas 1,
E.R._plas 1
|
(predict for NP_001328034.1)
|
|
cyto 6,
chlo 1,
nucl 1
|
(predict for NP_001328035.1)
|
|
cyto 6,
E.R. 2,
nucl 1,
E.R._plas 1
|
(predict for NP_001328036.1)
|
|
chlo 5,
chlo_mito 3,
cyto 2,
plas 1,
extr 1,
vacu 1
|
(predict for NP_195059.3)
|
|
Subcellular
localization
TargetP |
|
scret 7
|
(predict for NP_001154284.1)
|
|
other 6
|
(predict for NP_001328034.1)
|
|
other 8
|
(predict for NP_001328035.1)
|
|
other 6
|
(predict for NP_001328036.1)
|
|
scret 7
|
(predict for NP_195059.3)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with PGSIP3 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 10.7 |
GAUT12 |
galacturonosyltransferase 12 |
[detail] |
835558 |
| 10.3 |
AT5G60720 |
electron transporter, putative (Protein of unknown function, DUF547) |
[detail] |
836193 |
| 10.2 |
IRX9 |
Nucleotide-diphospho-sugar transferases superfamily protein |
[detail] |
818285 |
| 8.1 |
GXMT1 |
glucuronoxylan 4-O-methyltransferase-like protein (DUF579) |
[detail] |
840272 |
| 7.4 |
AT1G03920 |
Protein kinase family protein |
[detail] |
839369 |
| 6.6 |
KNAT7 |
homeobox knotted-like protein |
[detail] |
842602 |
| 5.3 |
AT1G76250 |
transmembrane protein |
[detail] |
843958 |
| 4.5 |
AT1G19190 |
alpha/beta-Hydrolases superfamily protein |
[detail] |
838502 |
|
Coexpressed
gene list |
[Coexpressed gene list for PGSIP3]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
253380_at
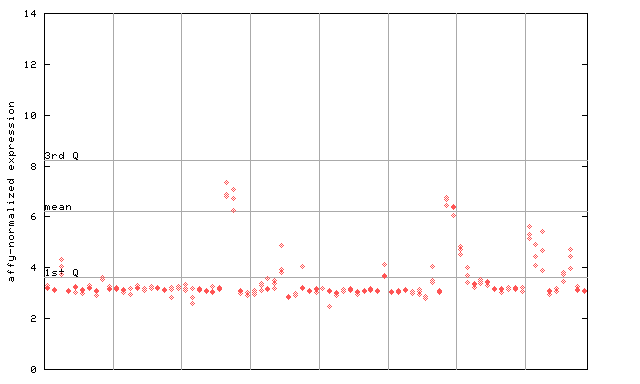
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
253380_at
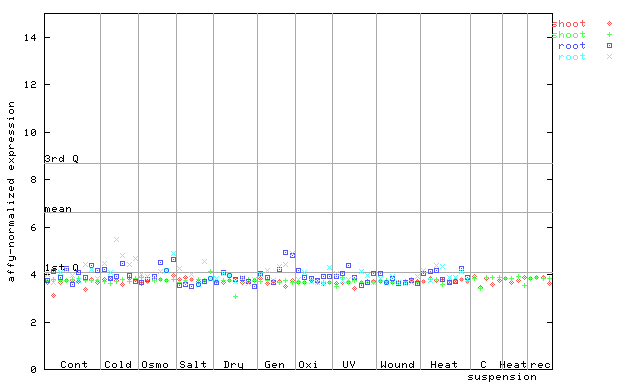
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
253380_at
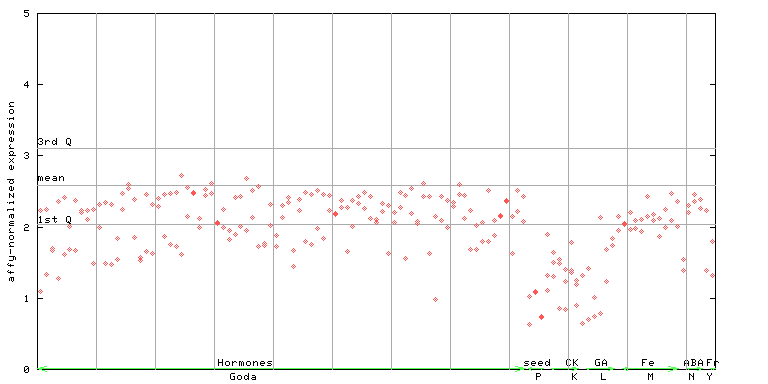
X axis is samples (xls file), and Y axis is log-expression.
|













