[←][→] ath
| functional annotation | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | phosphoserine aminotransferase |

 Plant GARDEN Plant GARDEN JBrowse
Plant GARDEN Plant GARDEN JBrowse
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | ath00260 [list] [network] Glycine, serine and threonine metabolism (70 genes) |  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00270 [list] [network] Cysteine and methionine metabolism (124 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath00750 [list] [network] Vitamin B6 metabolism (15 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath01200 [list] [network] Carbon metabolism (273 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath01230 [list] [network] Biosynthesis of amino acids (244 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ath01240 [list] [network] Biosynthesis of cofactors (236 genes) |  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_195288.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_195288.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] AT2G17630 (ath) LOC7492121 (ppo) LOC9270715 (osa) LOC11443166 (mtr) LOC100779355 (gma) LOC100795286 (gma) LOC100798600 (gma) LOC101254553 (sly) LOC103834946 (bra) LOC103837814 (bra) LOC123084884 (tae) LOC123093205 (tae) LOC123098468 (tae) LOC123449487 (hvu) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for PSAT] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
253162_at
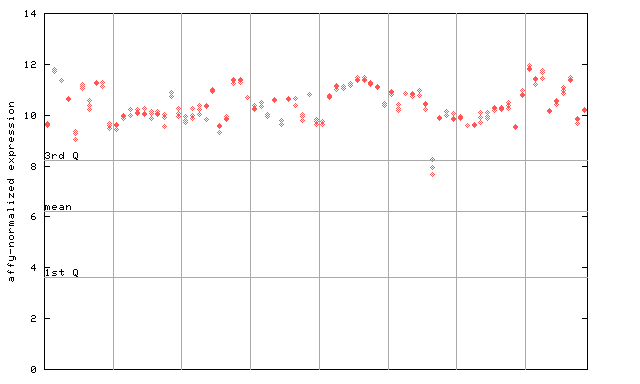
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
253162_at
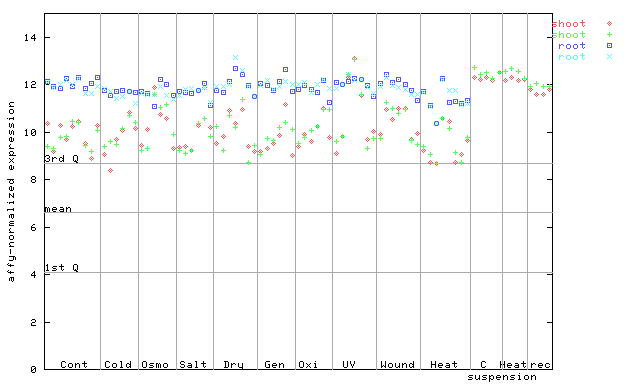
X axis is samples (pdf file), and Y axis is log2-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
253162_at
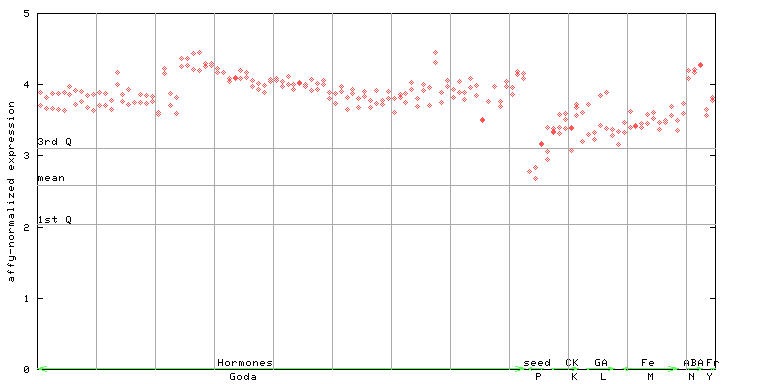
X axis is samples (xls file), and Y axis is log-expression. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 829715 |


|
| Refseq ID (protein) | NP_195288.1 |  |
The preparation time of this page was 0.2 [sec].






