| functional annotation |
| Function |
Protein kinase superfamily protein |


|
| GO BP |
|
GO:2000035 [list] [network] regulation of stem cell division
|
(1 genes)
|
IMP
|
 |
|
GO:2000069 [list] [network] regulation of post-embryonic root development
|
(13 genes)
|
IMP
|
 |
|
GO:0009750 [list] [network] response to fructose
|
(17 genes)
|
IMP
|
 |
|
GO:0010105 [list] [network] negative regulation of ethylene-activated signaling pathway
|
(17 genes)
|
TAS
|
 |
|
GO:0009686 [list] [network] gibberellin biosynthetic process
|
(26 genes)
|
IMP
|
 |
|
GO:0048510 [list] [network] regulation of timing of transition from vegetative to reproductive phase
|
(36 genes)
|
IMP
|
 |
|
GO:0010182 [list] [network] sugar mediated signaling pathway
|
(37 genes)
|
TAS
|
 |
|
GO:0009744 [list] [network] response to sucrose
|
(68 genes)
|
IMP
|
 |
|
GO:0046777 [list] [network] protein autophosphorylation
|
(173 genes)
|
IDA
|
 |
|
GO:0009873 [list] [network] ethylene-activated signaling pathway
|
(191 genes)
|
IEA
|
 |
|
GO:0001666 [list] [network] response to hypoxia
|
(263 genes)
|
IMP
|
 |
|
GO:0009723 [list] [network] response to ethylene
|
(297 genes)
|
IMP
|
 |
|
| GO CC |
|
GO:0005789 [list] [network] endoplasmic reticulum membrane
|
(466 genes)
|
IDA
|
 |
|
GO:0005634 [list] [network] nucleus
|
(10793 genes)
|
ISM
|
 |
|
| GO MF |
|
GO:0004712 [list] [network] protein serine/threonine/tyrosine kinase activity
|
(63 genes)
|
ISS
|
 |
|
GO:0004674 [list] [network] protein serine/threonine kinase activity
|
(801 genes)
|
IBA
IDA
|
 |
|
GO:0005524 [list] [network] ATP binding
|
(2003 genes)
|
IEA
|
 |
|
GO:0005515 [list] [network] protein binding
|
(4605 genes)
|
IPI
|
 |
|
| KEGG |
ath04016 [list] [network] MAPK signaling pathway - plant (134 genes) |
 |
| ath04075 [list] [network] Plant hormone signal transduction (273 genes) |
 |
| Protein |
NP_195993.1
NP_850760.1
|
| BLAST |
NP_195993.1
NP_850760.1
|
| Orthologous |
[Ortholog page]
CTR3 (sly)
CTR4 (sly)
CTR1 (sly)
LOC4329527 (osa)
LOC9270313 (osa)
LOC11423951 (mtr)
LOC25483894 (mtr)
LOC100191496 (zma)
LOC100240856 (vvi)
LOC100264370 (vvi)
LOC100775768 (gma)
LOC100787894 (gma)
LOC100800648 (gma)
LOC100807496 (gma)
LOC100810555 (gma)
CTR1 (bra)
LOC103855536 (bra)
|
Subcellular
localization
wolf |
|
chlo 6,
mito 2,
nucl 1,
cyto 1,
cyto_nucl 1
|
(predict for NP_195993.1)
|
|
chlo 6,
mito 2,
nucl 1,
cyto 1,
cyto_nucl 1
|
(predict for NP_850760.1)
|
|
Subcellular
localization
TargetP |
|
other 6,
mito 3
|
(predict for NP_195993.1)
|
|
other 6,
mito 3
|
(predict for NP_850760.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04016 |
MAPK signaling pathway - plant |
9 |
 |
| ath04075 |
Plant hormone signal transduction |
8 |
 |
Genes directly connected with CTR1 on the network
| coex z* |
Locus |
Function* |
Coexpression
detail |
Entrez Gene ID* |
| 8.2 |
EBF2 |
EIN3-binding F box protein 2 |
[detail] |
832607 |
| 8.0 |
ERS1 |
ethylene response sensor 1 |
[detail] |
818693 |
| 6.3 |
ETR2 |
Signal transduction histidine kinase, hybrid-type, ethylene sensor |
[detail] |
821891 |
| 6.2 |
AT4G38520 |
Protein phosphatase 2C family protein |
[detail] |
830009 |
| 5.3 |
CLT3 |
CRT (chloroquine-resistance transporter)-like transporter 3 |
[detail] |
831090 |
| 4.9 |
DGK5 |
diacylglycerol kinase 5 |
[detail] |
816624 |
|
Coexpressed
gene list |
[Coexpressed gene list for CTR1]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
250911_at
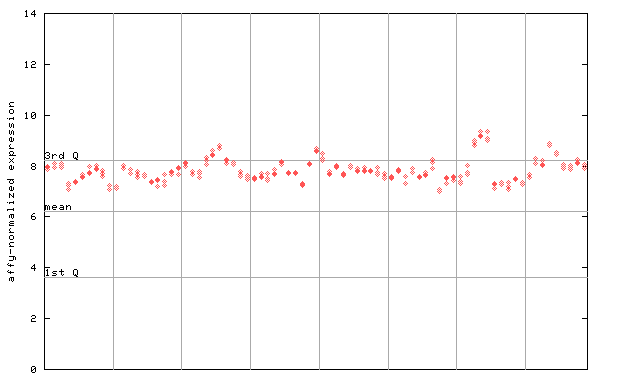
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
250911_at
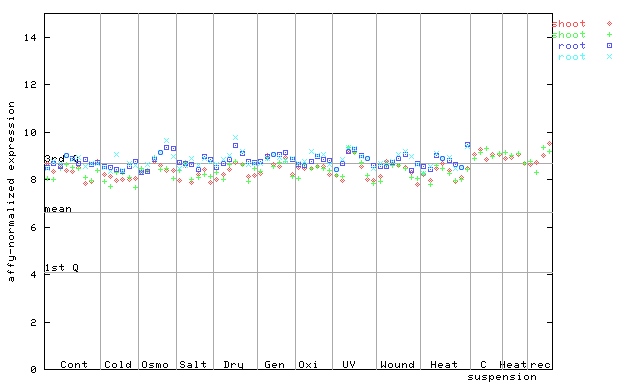
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
250911_at
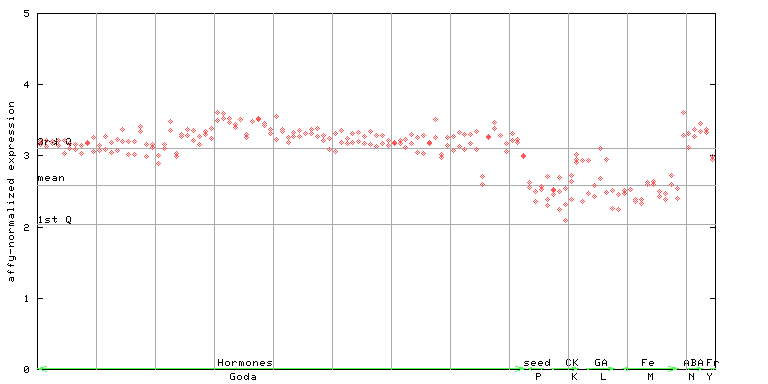
X axis is samples (xls file), and Y axis is log-expression.
|














