| functional annotation |
| Function |
PsbP domain protein (Mog1/PsbP/DUF1795-like photosystem II reaction center PsbP family protein) |




|
| GO BP |
|
GO:0015979 [list] [network] photosynthesis
|
(167 genes)
|
IEA
|
 |
|
GO:0044262 [list] [network] cellular carbohydrate metabolic process
|
(356 genes)
|
IEA
|
 |
|
GO:0009793 [list] [network] embryo development ending in seed dormancy
|
(697 genes)
|
IEA
|
 |
|
GO:0009266 [list] [network] response to temperature stimulus
|
(843 genes)
|
IEA
|
 |
|
| GO CC |
|
GO:0009654 [list] [network] photosystem II oxygen evolving complex
|
(19 genes)
|
IEA
|
 |
|
GO:0019898 [list] [network] extrinsic component of membrane
|
(46 genes)
|
IEA
|
 |
|
GO:0031977 [list] [network] thylakoid lumen
|
(67 genes)
|
HDA
|
 |
|
GO:0009535 [list] [network] chloroplast thylakoid membrane
|
(315 genes)
|
HDA
|
 |
|
GO:0009579 [list] [network] thylakoid
|
(509 genes)
|
HDA
|
 |
|
GO:0005886 [list] [network] plasma membrane
|
(2529 genes)
|
HDA
|
 |
|
GO:0005739 [list] [network] mitochondrion
|
(4228 genes)
|
ISM
|
 |
|
GO:0009507 [list] [network] chloroplast
|
(5004 genes)
|
HDA
ISM
|
 |
|
| GO MF |
|
GO:0005509 [list] [network] calcium ion binding
|
(132 genes)
|
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001331121.1
NP_196706.2
|
| BLAST |
NP_001331121.1
NP_196706.2
|
| Orthologous |
[Ortholog page]
LOC4326610 (osa)
LOC7471559 (ppo)
LOC11423366 (mtr)
LOC100776365 (gma)
LOC100809891 (gma)
LOC101249944 (sly)
LOC103846717 (bra)
LOC123149440 (tae)
LOC123407647 (hvu)
|
Subcellular
localization
wolf |
|
nucl 9,
chlo 1,
mito 1,
chlo_mito 1
|
(predict for NP_001331121.1)
|
|
chlo 6,
mito 4
|
(predict for NP_196706.2)
|
|
Subcellular
localization
TargetP |
|
chlo 7
|
(predict for NP_001331121.1)
|
|
chlo 8,
mito 3
|
(predict for NP_196706.2)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03010 |
Ribosome |
3 |
 |
Genes directly connected with PPD5 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 9.9 |
AT2G24060 |
Translation initiation factor 3 protein |
[detail] |
816940 |
| 9.8 |
AT3G13180 |
NOL1/NOP2/sun family protein / antitermination NusB domain-containing protein |
[detail] |
820508 |
| 9.6 |
AT4G17560 |
Ribosomal protein L19 family protein |
[detail] |
827471 |
| 9.3 |
AT3G47650 |
DnaJ/Hsp40 cysteine-rich domain superfamily protein |
[detail] |
823919 |
| 9.2 |
PDF1B |
peptide deformylase 1B |
[detail] |
831318 |
| 8.4 |
AT3G59980 |
Nucleic acid-binding, OB-fold-like protein |
[detail] |
825168 |
| 7.0 |
AT2G33180 |
uncharacterized protein |
[detail] |
817879 |
| 6.0 |
EMB3123 |
PPR containing protein |
[detail] |
822397 |
| 5.5 |
AT1G53120 |
RNA-binding S4 domain-containing protein |
[detail] |
841746 |
|
Coexpressed
gene list |
[Coexpressed gene list for PPD5]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
250371_at
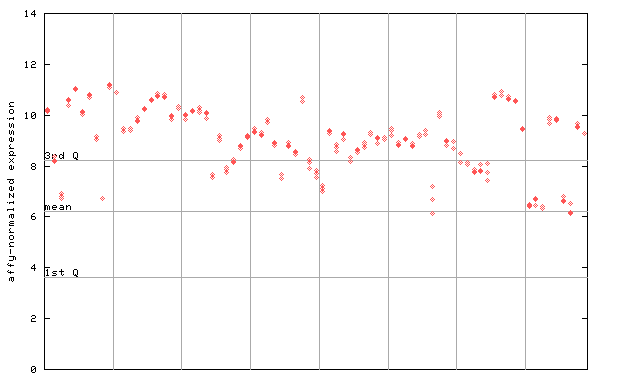
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
250371_at
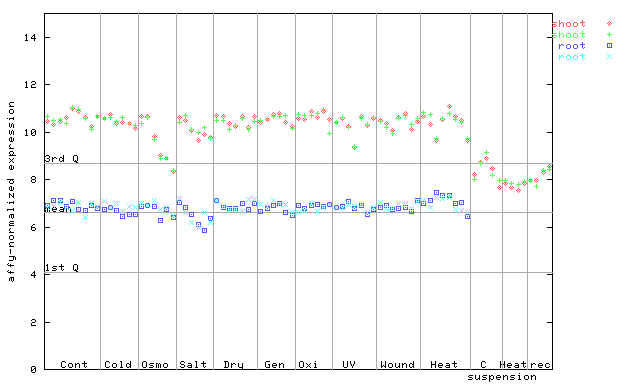
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
250371_at
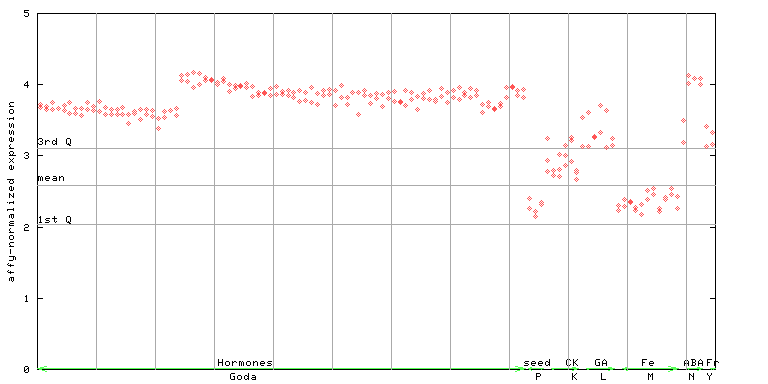
X axis is samples (xls file), and Y axis is log-expression.
|













