| functional annotation |
| Function |
delta subunit of Mt ATP synthase |




|
| GO BP |
|
| GO CC |
|
GO:0000275 [list] [network] mitochondrial proton-transporting ATP synthase complex, catalytic sector F(1)
|
(5 genes)
|
ISS
|
 |
|
GO:0005753 [list] [network] mitochondrial proton-transporting ATP synthase complex
|
(12 genes)
|
HDA
|
 |
|
GO:0005794 [list] [network] Golgi apparatus
|
(1182 genes)
|
RCA
|
 |
|
GO:0005739 [list] [network] mitochondrion
|
(4228 genes)
|
HDA
IDA
ISM
|
 |
|
GO:0009507 [list] [network] chloroplast
|
(5004 genes)
|
HDA
|
 |
|
GO:0009536 [list] [network] plastid
|
(5425 genes)
|
HDA
|
 |
|
| GO MF |
|
GO:0046933 [list] [network] proton-transporting ATP synthase activity, rotational mechanism
|
(8 genes)
|
ISS
|
 |
|
GO:0050897 [list] [network] cobalt ion binding
|
(44 genes)
|
HDA
|
 |
|
GO:0008270 [list] [network] zinc ion binding
|
(429 genes)
|
HDA
|
 |
|
| KEGG |
ath00190 [list] [network] Oxidative phosphorylation (170 genes) |
 |
| Protein |
NP_001031875.1
NP_196849.1
|
| BLAST |
NP_001031875.1
NP_196849.1
|
| Orthologous |
[Ortholog page]
LOC542932 (tae)
LOC4341660 (osa)
LOC7467328 (ppo)
LOC25488771 (mtr)
LOC100499796 (gma)
LOC100790990 (gma)
LOC101244808 (sly)
LOC103846573 (bra)
LOC103850871 (bra)
LOC103856017 (bra)
LOC123128151 (tae)
LOC123137946 (tae)
LOC123402581 (hvu)
|
Subcellular
localization
wolf |
|
mito 6,
chlo 4
|
(predict for NP_001031875.1)
|
|
mito 5,
chlo 4,
cyto 1,
plas 1,
cyto_plas 1
|
(predict for NP_196849.1)
|
|
Subcellular
localization
TargetP |
|
mito 4,
chlo 3
|
(predict for NP_001031875.1)
|
|
mito 4
|
(predict for NP_196849.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath00190 |
Oxidative phosphorylation |
12 |
 |
Genes directly connected with ATP5 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 16.1 |
ATP3 |
gamma subunit of Mt ATP synthase |
[detail] |
817866 |
| 16.0 |
GAMMA CA3 |
gamma carbonic anhydrase 3 |
[detail] |
836783 |
| 15.9 |
MGP1 |
MALE GAMETOPHYTE DEFECTIVE 1 |
[detail] |
816723 |
| 12.9 |
GAMMA CA2 |
gamma carbonic anhydrase 2 |
[detail] |
841129 |
| 12.4 |
ATPQ |
ATP synthase D chain |
[detail] |
824395 |
| 11.8 |
AT4G02580 |
NADH-ubiquinone oxidoreductase 24 kDa subunit |
[detail] |
828130 |
| 11.3 |
AT5G47030 |
ATPase, F1 complex, delta/epsilon subunit |
[detail] |
834749 |
| 11.0 |
AT5G20080 |
FAD/NAD(P)-binding oxidoreductase |
[detail] |
832130 |
| 10.4 |
COX6B |
cytochrome C oxidase 6B |
[detail] |
838851 |
| 6.9 |
FLDH |
NAD(P)-binding Rossmann-fold superfamily protein |
[detail] |
829473 |
| 6.9 |
AT2G40800 |
import inner membrane translocase subunit |
[detail] |
818677 |
| 5.5 |
ACR12 |
ACT domain-containing protein |
[detail] |
830352 |
| 5.2 |
bHLH115 |
basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
[detail] |
841529 |
|
Coexpressed
gene list |
[Coexpressed gene list for ATP5]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
250236_at
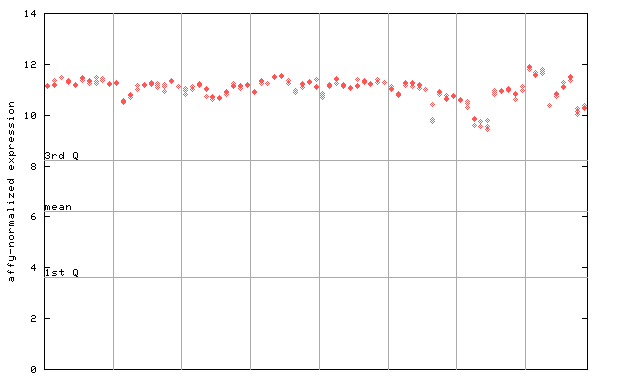
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
250236_at
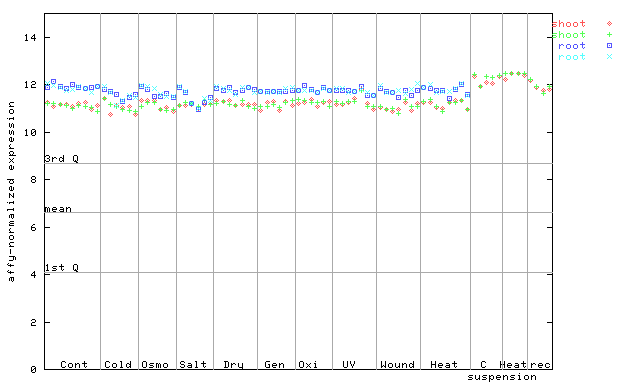
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
250236_at
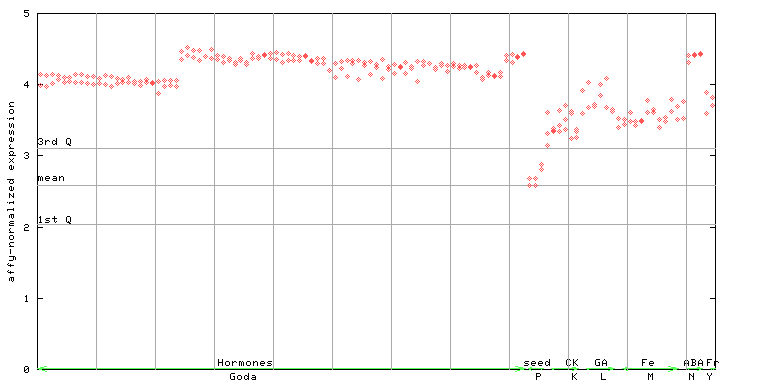
X axis is samples (xls file), and Y axis is log-expression.
|














