| functional annotation |
| Function |
double-stranded DNA binding protein |




|
| GO BP |
|
| GO CC |
|
GO:0009330 [list] [network] DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) complex
|
(3 genes)
|
IEA
IPI
|
 |
|
GO:0005634 [list] [network] nucleus
|
(10305 genes)
|
IDA
|
 |
|
| GO MF |
|
GO:0003690 [list] [network] double-stranded DNA binding
|
(789 genes)
|
IDA
IEA
|
 |
|
| KEGG |
|
|
| Protein |
NP_001154735.1
NP_001154736.1
NP_001154737.1
NP_001154738.1
NP_001154739.1
NP_001330963.1
NP_001330964.1
NP_001330965.1
NP_001330966.1
NP_001330967.1
NP_001330968.1
NP_197851.5
|
| BLAST |
NP_001154735.1
NP_001154736.1
NP_001154737.1
NP_001154738.1
NP_001154739.1
NP_001330963.1
NP_001330964.1
NP_001330965.1
NP_001330966.1
NP_001330967.1
NP_001330968.1
NP_197851.5
|
| Orthologous |
[Ortholog page]
LOC4328289 (osa)
LOC4328294 (osa)
LOC7486704 (ppo)
LOC11443488 (mtr)
LOC100795660 (gma)
LOC100812582 (gma)
LOC101256852 (sly)
LOC103837066 (bra)
LOC103874327 (bra)
LOC123132237 (tae)
LOC123136281 (tae)
LOC123143785 (tae)
LOC123401307 (hvu)
|
Subcellular
localization
wolf |
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_001154735.1)
|
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_001154736.1)
|
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_001154737.1)
|
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_001154738.1)
|
|
nucl 9,
cyto_nucl 5
|
(predict for NP_001154739.1)
|
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_001330963.1)
|
|
nucl 7,
cyto_nucl 4,
cyto 1
|
(predict for NP_001330964.1)
|
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_001330965.1)
|
|
nucl 7,
cyto_nucl 4,
cyto 1
|
(predict for NP_001330966.1)
|
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_001330967.1)
|
|
nucl 7,
cyto_nucl 4,
cyto 1
|
(predict for NP_001330968.1)
|
|
nucl 8,
cyto_nucl 5,
cyto 1
|
(predict for NP_197851.5)
|
|
Subcellular
localization
TargetP |
|
other 5,
chlo 3
|
(predict for NP_001154735.1)
|
|
other 5,
chlo 3
|
(predict for NP_001154736.1)
|
|
other 5,
chlo 3
|
(predict for NP_001154737.1)
|
|
other 5,
chlo 3
|
(predict for NP_001154738.1)
|
|
other 6,
chlo 3
|
(predict for NP_001154739.1)
|
|
other 5,
chlo 3
|
(predict for NP_001330963.1)
|
|
other 5,
chlo 3
|
(predict for NP_001330964.1)
|
|
other 5,
chlo 3
|
(predict for NP_001330965.1)
|
|
other 5,
chlo 3
|
(predict for NP_001330966.1)
|
|
other 5,
chlo 3
|
(predict for NP_001330967.1)
|
|
other 5,
chlo 3
|
(predict for NP_001330968.1)
|
|
other 5,
chlo 4
|
(predict for NP_197851.5)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath03030 |
DNA replication |
6 |
 |
| ath03410 |
Base excision repair |
5 |
 |
| ath03430 |
Mismatch repair |
4 |
 |
| ath03420 |
Nucleotide excision repair |
4 |
 |
| ath03440 |
Homologous recombination |
3 |
 |
Genes directly connected with BIN4 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 7.0 |
POLD3 |
DNA-directed DNA polymerase |
[detail] |
844201 |
| 6.6 |
MEI1 |
transcription coactivator |
[detail] |
844068 |
| 5.4 |
MIM |
P-loop containing nucleoside triphosphate hydrolases superfamily protein |
[detail] |
836267 |
|
Coexpressed
gene list |
[Coexpressed gene list for BIN4]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
249749_at
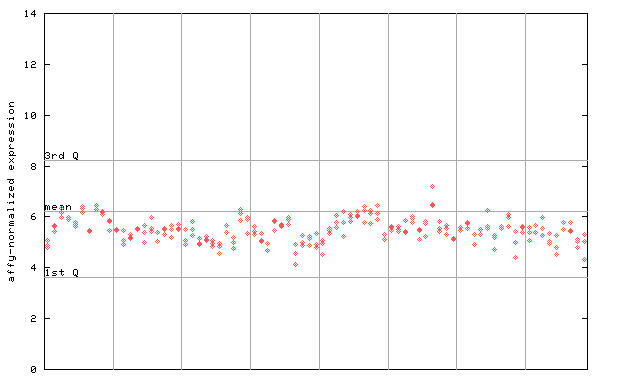
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
249749_at
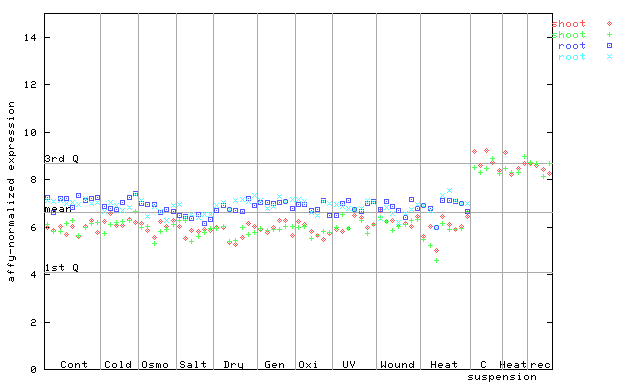
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
249749_at
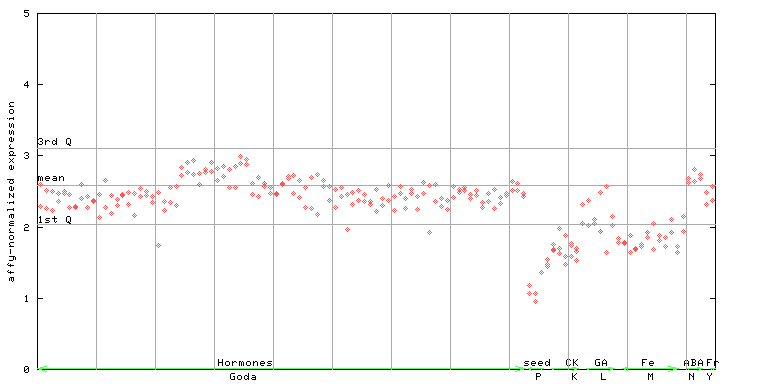
X axis is samples (xls file), and Y axis is log-expression.
|













