| functional annotation |
| Function |
jasmonate-zim-domain protein 5 |




|
| GO BP |
|
| GO CC |
|
GO:0009507 [list] [network] chloroplast
|
(5004 genes)
|
ISM
|
 |
|
GO:0005634 [list] [network] nucleus
|
(10305 genes)
|
ISM
|
 |
|
| GO MF |
|
| KEGG |
ath04075 [list] [network] Plant hormone signal transduction (291 genes) |
 |
| Protein |
NP_001320904.1
NP_001320905.1
NP_564019.1
|
| BLAST |
NP_001320904.1
NP_001320905.1
NP_564019.1
|
| Orthologous |
[Ortholog page]
LOC103835959 (bra)
LOC103842752 (bra)
LOC103872642 (bra)
|
Subcellular
localization
wolf |
|
nucl 8,
nucl_plas 4,
chlo 1,
extr 1
|
(predict for NP_001320904.1)
|
|
mito 4,
nucl 3,
cyto_nucl 2,
cyto_mito 2
|
(predict for NP_001320905.1)
|
|
nucl 9,
chlo 1,
extr 1
|
(predict for NP_564019.1)
|
|
Subcellular
localization
TargetP |
|
other 4,
chlo 3
|
(predict for NP_001320904.1)
|
|
scret 8,
mito 3
|
(predict for NP_001320905.1)
|
|
other 4,
chlo 3
|
(predict for NP_564019.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
| KEGG* ID |
Title |
#genes |
Link to the KEGG* map
(Multiple genes) |
| ath04075 |
Plant hormone signal transduction |
6 |
 |
| ath00592 |
alpha-Linolenic acid metabolism |
5 |
 |
| ath00591 |
Linoleic acid metabolism |
2 |
 |
Genes directly connected with JAZ5 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 11.3 |
JAZ10 |
jasmonate-zim-domain protein 10 |
[detail] |
831162 |
| 11.3 |
JAZ1 |
jasmonate-zim-domain protein 1 |
[detail] |
838501 |
| 11.1 |
TIFY10B |
TIFY domain/Divergent CCT motif family protein |
[detail] |
843834 |
| 10.5 |
JAZ6 |
jasmonate-zim-domain protein 6 |
[detail] |
843577 |
| 10.5 |
OPR3 |
oxophytodienoate-reductase 3 |
[detail] |
815160 |
| 10.1 |
LOX4 |
PLAT/LH2 domain-containing lipoxygenase family protein |
[detail] |
843584 |
| 10.1 |
JAZ8 |
jasmonate-zim-domain protein 8 |
[detail] |
839893 |
| 10.0 |
LOX3 |
lipoxygenase 3 |
[detail] |
838314 |
| 9.6 |
OPCL1 |
OPC-8:0 CoA ligase1 |
[detail] |
838639 |
| 8.5 |
AOC3 |
allene oxide cyclase 3 |
[detail] |
822169 |
| 7.3 |
MAPKKK21 |
mitogen-activated protein kinase kinase kinase 21 |
[detail] |
3770588 |
| 7.0 |
AT2G42760 |
DUF1685 family protein |
[detail] |
818876 |
| 7.0 |
AIB |
ABA-inducible BHLH-type transcription factor |
[detail] |
819262 |
| 6.0 |
GRX480 |
Thioredoxin superfamily protein |
[detail] |
839748 |
| 5.4 |
AT5G14700 |
NAD(P)-binding Rossmann-fold superfamily protein |
[detail] |
831322 |
| 5.4 |
AT1G61610 |
S-locus lectin protein kinase family protein |
[detail] |
842457 |
| 5.0 |
BHLH92 |
basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
[detail] |
834385 |
|
Coexpressed
gene list |
[Coexpressed gene list for JAZ5]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
261033_at
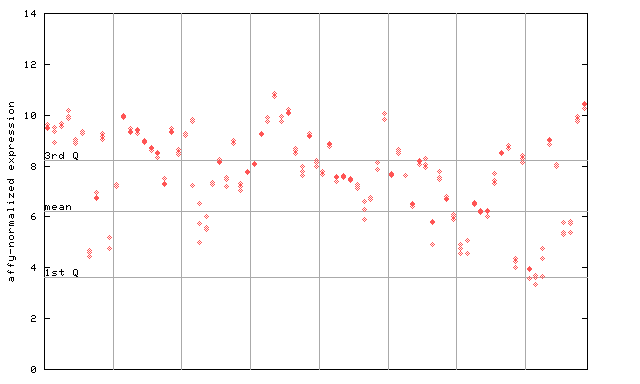
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
261033_at
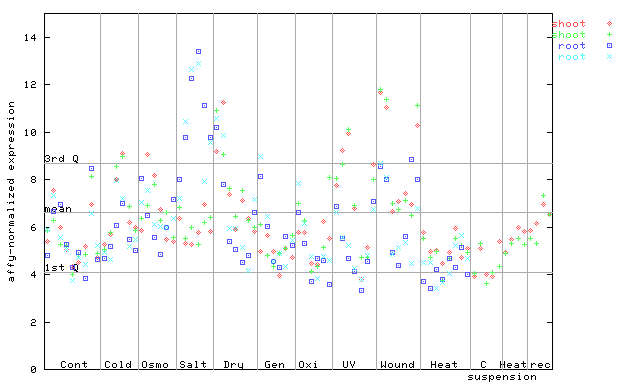
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
261033_at
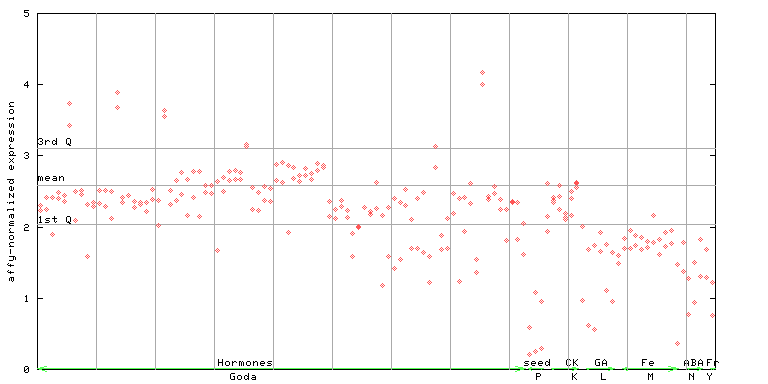
X axis is samples (xls file), and Y axis is log-expression.
|

















