[←][→] ath
| functional annotation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Function | Leucine-rich repeat protein kinase family protein |




|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO BP |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO CC |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO MF |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein | NP_175591.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BLAST | NP_175591.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Orthologous | [Ortholog page] AT2G04300 (ath) AT2G14440 (ath) AT2G14510 (ath) FRK1 (ath) AT2G19210 (ath) AT2G19230 (ath) AT2G28960 (ath) AT2G28970 (ath) AT2G28990 (ath) AT2G29000 (ath) AT3G21340 (ath) MEE39 (ath) AT3G46340 (ath) AT3G46370 (ath) AT3G46400 (ath) AT3G46420 (ath) AT4G20450 (ath) RHS16 (ath) AT4G29450 (ath) AT4G29990 (ath) AT5G16900 (ath) AT5G59650 (ath) AT5G59670 (ath) AT5G59680 (ath) AT1G05700 (ath) AT1G07550 (ath) AT1G07560 (ath) AT1G49100 (ath) AT1G51790 (ath) AT1G51805 (ath) AT1G51810 (ath) AT1G51820 (ath) AT1G51830 (ath) AT1G51850 (ath) AT1G51860 (ath) AT1G51870 (ath) RHS6 (ath) AT1G51890 (ath) AT1G51910 (ath) LOC4328315 (osa) LOC4338207 (osa) LOC4339370 (osa) LOC4339371 (osa) LOC4339374 (osa) LOC4339376 (osa) LOC4339378 (osa) LOC4346819 (osa) LOC4346820 (osa) LOC4346821 (osa) LOC4346822 (osa) LOC4346823 (osa) LOC4346825 (osa) LOC4346833 (osa) LOC4346837 (osa) LOC4346841 (osa) LOC7494617 (ppo) LOC9266163 (osa) LOC9267814 (osa) LOC9267979 (osa) LOC9272354 (osa) LOC11421236 (mtr) LOC11422007 (mtr) LOC11425516 (mtr) LOC11429446 (mtr) LOC11433438 (mtr) LOC11434037 (mtr) LOC11437028 (mtr) LOC18108501 (ppo) LOC18108503 (ppo) LOC18108531 (ppo) LOC18110286 (ppo) LOC18110288 (ppo) LOC18110448 (ppo) LOC18110542 (ppo) LOC18110545 (ppo) LOC18110550 (ppo) LOC25497008 (mtr) LOC25500355 (mtr) LOC25500358 (mtr) LOC25500359 (mtr) LOC25500361 (mtr) LOC25500364 (mtr) LOC25500366 (mtr) LOC25500367 (mtr) LOC25500376 (mtr) LOC25500764 (mtr) LOC25502465 (mtr) SIK2 (gma) LOC100775741 (gma) LOC100776284 (gma) LOC100776818 (gma) LOC100778537 (gma) LOC100779102 (gma) LOC100779474 (gma) LOC100779617 (gma) LOC100781084 (gma) LOC100781785 (gma) LOC100785345 (gma) LOC100791717 (gma) LOC100792239 (gma) LOC100793137 (gma) LOC100794504 (gma) LOC100795183 (gma) LOC100818475 (gma) LOC101257947 (sly) LOC101262429 (sly) LOC101266705 (sly) LOC102662730 (gma) LOC102663143 (gma) LOC103829493 (bra) LOC103829497 (bra) LOC103836435 (bra) LOC103843445 (bra) LOC103844100 (bra) LOC103845385 (bra) LOC103850089 (bra) LOC103851563 (bra) LOC103851564 (bra) LOC103851565 (bra) LOC103851763 (bra) LOC103853036 (bra) LOC103853548 (bra) LOC103856616 (bra) LOC103859903 (bra) LOC103861877 (bra) LOC103864933 (bra) LOC103864934 (bra) LOC103868533 (bra) LOC103868536 (bra) LOC103868540 (bra) LOC103868542 (bra) LOC103868543 (bra) LOC103868545 (bra) LOC103868632 (bra) LOC103869180 (bra) LOC103871250 (bra) LOC103871251 (bra) LOC103871253 (bra) LOC103871255 (bra) LOC103873634 (bra) LOC106794186 (gma) LOC106799536 (gma) LOC107275782 (osa) LOC107276230 (osa) LOC107276752 (osa) LOC107278492 (osa) LOC107278845 (osa) LOC112325225 (ppo) LOC112416101 (mtr) LOC112417465 (mtr) LOC112936039 (osa) LOC117125683 (bra) LOC120575758 (mtr) LOC123043522 (tae) LOC123044006 (tae) LOC123044087 (tae) LOC123044243 (tae) LOC123054720 (tae) LOC123063692 (tae) LOC123067573 (tae) LOC123079142 (tae) LOC123080188 (tae) LOC123080196 (tae) LOC123080205 (tae) LOC123080238 (tae) LOC123083057 (tae) LOC123084992 (tae) LOC123085003 (tae) LOC123086249 (tae) LOC123094168 (tae) LOC123094169 (tae) LOC123096526 (tae) LOC123096537 (tae) LOC123096545 (tae) LOC123096554 (tae) LOC123096599 (tae) LOC123096617 (tae) LOC123096813 (tae) LOC123097489 (tae) LOC123103544 (tae) LOC123106972 (tae) LOC123114350 (tae) LOC123114773 (tae) LOC123116086 (tae) LOC123120524 (tae) LOC123124720 (tae) LOC123126207 (tae) LOC123132270 (tae) LOC123132271 (tae) LOC123132273 (tae) LOC123136316 (tae) LOC123136318 (tae) LOC123136319 (tae) LOC123138953 (tae) LOC123140966 (tae) LOC123143830 (tae) LOC123143832 (tae) LOC123143833 (tae) LOC123143893 (tae) LOC123157067 (tae) LOC123160036 (tae) LOC123160179 (tae) LOC123160205 (tae) LOC123160231 (tae) LOC123162979 (tae) LOC123162990 (tae) LOC123173379 (tae) LOC123173395 (tae) LOC123173449 (tae) LOC123179719 (tae) LOC123179738 (tae) LOC123179780 (tae) LOC123180475 (tae) LOC123182688 (tae) LOC123182689 (tae) LOC123395140 (hvu) LOC123398290 (hvu) LOC123401280 (hvu) LOC123401354 (hvu) LOC123401635 (hvu) LOC123404089 (hvu) LOC123404264 (hvu) LOC123404421 (hvu) LOC123404434 (hvu) LOC123404717 (hvu) LOC123408210 (hvu) LOC123416831 (hvu) LOC123416847 (hvu) LOC123416895 (hvu) LOC123440187 (hvu) LOC123442690 (hvu) LOC123448982 (hvu) LOC123450805 (hvu) LOC123450806 (hvu) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization wolf |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Subcellular localization TargetP |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene coexpression | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Network*for coexpressed genes |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coexpressed gene list |
[Coexpressed gene list for IOS1] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene expression | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| All samples | [Expression pattern for all samples] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Development) |
256169_at
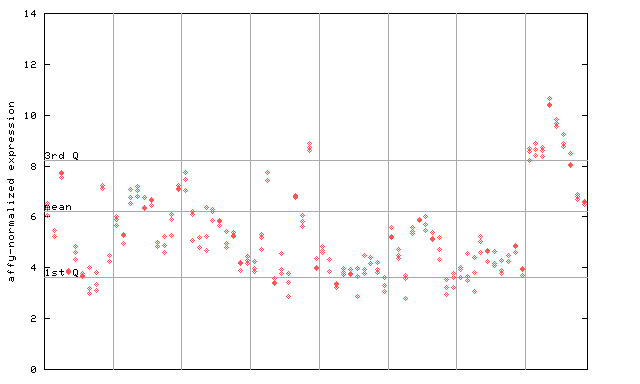
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Stress) |
256169_at
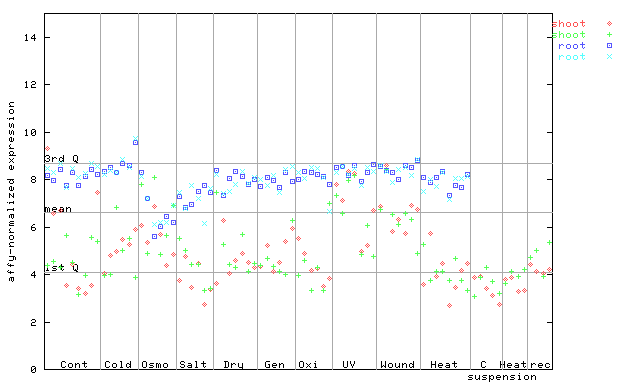
X axis is samples (pdf file), and Y axis is log2-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AtGenExpress* (Hormone) |
256169_at
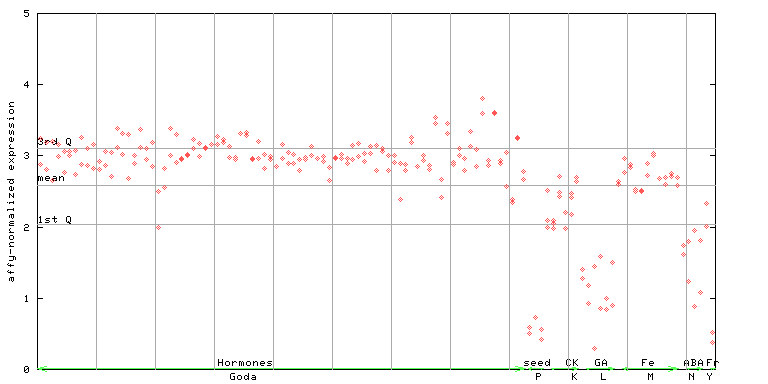
X axis is samples (xls file), and Y axis is log-expression. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Link to other DBs | ||
| Entrez Gene ID | 841606 |


|
| Refseq ID (protein) | NP_175591.1 |  |
The preparation time of this page was 0.1 [sec].

