| functional annotation |
| Function |
beta glucosidase 18 |




|
| GO BP |
|
GO:0080119 [list] [network] ER body organization
|
(6 genes)
|
IMP
|
 |
|
GO:0030104 [list] [network] water homeostasis
|
(15 genes)
|
IMP
|
 |
|
GO:0009687 [list] [network] abscisic acid metabolic process
|
(21 genes)
|
IDA
|
 |
|
GO:0051258 [list] [network] protein polymerization
|
(27 genes)
|
IDA
|
 |
|
GO:0009625 [list] [network] response to insect
|
(33 genes)
|
IEP
|
 |
|
GO:0009789 [list] [network] positive regulation of abscisic acid-activated signaling pathway
|
(53 genes)
|
IMP
|
 |
|
GO:0010119 [list] [network] regulation of stomatal movement
|
(105 genes)
|
IMP
|
 |
|
GO:0009651 [list] [network] response to salt stress
|
(607 genes)
|
IEP
|
 |
|
GO:0050832 [list] [network] defense response to fungus
|
(705 genes)
|
IEP
|
 |
|
GO:0009414 [list] [network] response to water deprivation
|
(1006 genes)
|
IEP
|
 |
|
GO:0009737 [list] [network] response to abscisic acid
|
(1086 genes)
|
IEP
|
 |
|
| GO CC |
|
| GO MF |
|
GO:0051993 [list] [network] abscisic acid glucose ester beta-glucosidase activity
|
(1 genes)
|
IDA
|
 |
|
GO:0008422 [list] [network] beta-glucosidase activity
|
(65 genes)
|
IMP
|
 |
|
GO:0042802 [list] [network] identical protein binding
|
(404 genes)
|
IPI
|
 |
|
| KEGG |
ath00906 [list] [network] Carotenoid biosynthesis (29 genes) |
 |
| Protein |
NP_001031175.1
NP_001185204.1
NP_175649.1
|
| BLAST |
NP_001031175.1
NP_001185204.1
NP_175649.1
|
| Orthologous |
[Ortholog page]
LOC103868505 (bra)
LOC103871211 (bra)
|
Subcellular
localization
wolf |
|
vacu 4,
extr 1,
E.R. 1,
chlo 1,
cyto 1,
pero 1,
cysk 1,
golg 1,
cyto_pero 1,
E.R._plas 1
|
(predict for NP_001031175.1)
|
|
vacu 4,
extr 1,
E.R. 1,
golg 1,
chlo 1,
pero 1,
golg_plas 1,
E.R._plas 1,
cyto_E.R. 1
|
(predict for NP_001185204.1)
|
|
vacu 4,
extr 1,
E.R. 1,
golg 1,
chlo 1,
pero 1,
golg_plas 1,
E.R._plas 1,
cyto_E.R. 1
|
(predict for NP_175649.1)
|
|
Subcellular
localization
TargetP |
|
scret 9
|
(predict for NP_001031175.1)
|
|
scret 9
|
(predict for NP_001185204.1)
|
|
scret 9
|
(predict for NP_175649.1)
|
|
| Gene coexpression |
Network*for
coexpressed
genes |
Genes directly connected with BGLU18 on the network
| coex z* |
Locus |
Function* |
CoexViewer |
Entrez Gene ID* |
| 22.0 |
TSA1 |
TSK-associating protein 1 |
[detail] |
841671 |
| 15.6 |
MBP1 |
myrosinase-binding protein 1 |
[detail] |
841633 |
| 15.1 |
MBP2 |
myrosinase-binding protein 2 |
[detail] |
841632 |
| 13.1 |
AT1G52000 |
Mannose-binding lectin superfamily protein |
[detail] |
841629 |
| 11.7 |
AT3G28220 |
TRAF-like family protein |
[detail] |
822448 |
| 10.4 |
JR1 |
Mannose-binding lectin superfamily protein |
[detail] |
820895 |
| 10.3 |
CSLA10 |
cellulose synthase-like A10 |
[detail] |
839019 |
| 9.2 |
PGL5 |
6-phosphogluconolactonase 5 |
[detail] |
832513 |
| 8.3 |
AT2G43530 |
scorpion toxin-like knottin superfamily protein |
[detail] |
818954 |
|
Coexpressed
gene list |
[Coexpressed gene list for BGLU18]
|
| Gene expression |
| All samples |
[Expression pattern for all samples]
|
AtGenExpress*
(Development) |
259640_at
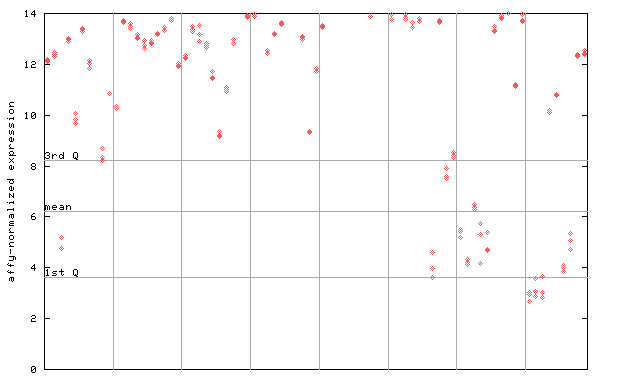
X axis is samples (pdf file), and Y axis is log2-expression.
"1st Q", "mean" and "3rd Q" indecate the values for all genes on a GeneChip. Q: quartile.
|
AtGenExpress*
(Stress) |
259640_at
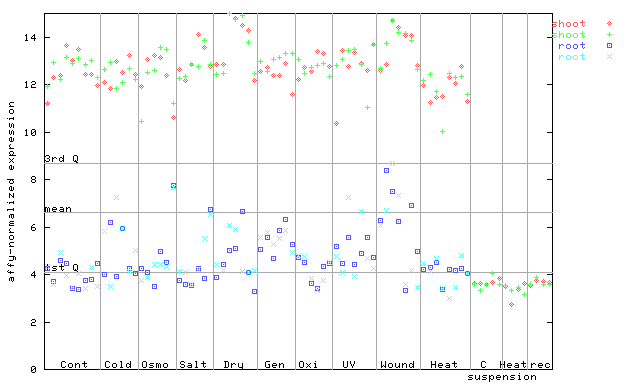
X axis is samples (pdf file), and Y axis is log2-expression.
|
AtGenExpress*
(Hormone) |
259640_at
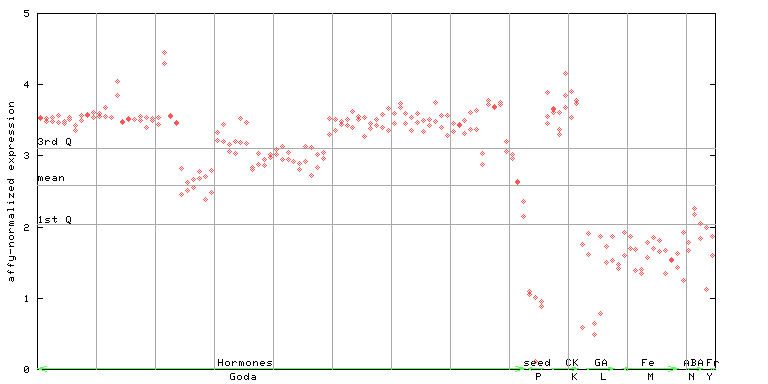
X axis is samples (xls file), and Y axis is log-expression.
|














